Abstract
The present study reports the mitochondrial genome of A. hastata (Oberthür, 1892), a little-known Aporia species endemic to the southern margin of the Hengduan Mountains in Yunnan Province. This genome is circular, 15,148 bp in length, and consists of 13 PCGs, 22 tRNAs, and two rRNAs. The Bayesian phylogenetic tree clusters A. hastata with other Aporia taxa inside tribe Pierini Duponchel, [1835]. The findings of this study add valuable new information to the genus Aporia and are beneficial to a better understanding of phylogeography of these butterflies.
1. Introduction
The genus Aporia Hübner, [1819] is a unique group of butterflies rich in regional endemic species as a result of radiative speciation (Kanoh et al. Citation2017), especially in the Hengduan Mountains of Southwest China. Aporia hastata (Oberthür, 1892) is a narrow-ranged species endemic to the upper Yangtze River watershed in northwestern Yunnan (Della Bruna et al. Citation2013).
Since its original description in 1892, only a limited number of specimens were available for taxonomic research, not to mention genetic studies. During a recent expedition to northwestern Yunnan, the authors collected specimens of A. hastata and successfully sequenced the mitochondrial genome from one sample. This is the first genetic data of this little-known regional endemic butterfly, which would decipher its phylogenetic position and benefit phylogeography (Wang et al. Citation2020).
2. Materials and methods
2.1. Sample collection and preservation
The utilized specimen was collected by netting from Fengyu (25.982567°N, 99.963927°E, 2,200 m), Eryuan County, Yunnan Province, China. Three legs on the same side were extracted and preserved in ethanol within 4 h after collection, the spread specimen () was deposited in the Zoological Museum (insect collection) of Yunnan University, Kunming, China (specimen number: YNU-LEP-PIE-2022HAS01, contact person: Shao-Ji Hu, [email protected]).
2.2. DNA extraction and sequencing
Genomic DNA extraction using all three legs with the TIANamp Genomic DNA Kit (TianGen Biotech Co., Ltd., Beijing, China), and examined with 1% agarose gel electrophoresis for quality control.
The PCR primers were designed using Primer 5 (http://www.premierbiosoft.com/) and combined for longer fragments in the PCR reaction (Table S1) using TaKaRa Ex Taq Kit (TaKaRa Biomedical Technology, Beijing, China) with default reaction system. The thermal profile of PCR consists of an initial denaturation at 97 °C for 3 min, followed by 35 cycles of denaturation at 97 °C for 15 s and annealing at 55 °C for 3 min, and then a final elongation at 72 °C for 5 min.
PCR products were examined using 1% agarose gel electrophoresis (Figure S1, supplementary package) and purified using the TIANgel Purification Kit (TianGen Biotech Co., Ltd.). The purified PCR products were sequenced in both directions with primers combinations for 10 shorter fragments (∼800 bp each) (Table S1, supplementary package) on an ABI 3730xl sequencer (Applied Biosystems, Foster City, CA, USA).
2.3. Genome assembling and phylogenetic analysis
Raw sequence fragments (supplementary package) were assembled using DNAStar (https://www.dnastar.com/) with A. bieti (Oberthür, 1884) (KX495165) as the reference genome (Cao et al. Citation2016). tRNAs and rRNAs were identified using MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013), while PCGs were identified by BLASTn of NCBI (https://blast.ncbi.nlm.nih.gov/). The genome structure was mapped using CGView (Grant & Stothard, Citation2008).
A Bayesian phylogenetic tree was reconstructed using PhyloSuite 1.2.2 (Zhang et al. Citation2020) for 1,000,000 generations, with the optimal GTR + F + I + G4 model identified by ModelFinder (Kalyaanamoorthy et al. Citation2017). Twenty-five published sequences of Pierinae species were used as ingroups, while Catopsilia pomona (Linnaeus, 1758) (JX274649) (Pieridae: Coliadinae) and Leptidea morsei (Fenton, 1882) (JX274648) (Pieridae: Dismorphinae) was chosen as the outgroups. Currently accepted species names were adopted herein (Back, Citation2020; Cao et al. Citation2016; Della Bruna et al. Citation2013; van Gasse, Citation2021).
3. Results
3.1. Characteristics of A. hastata mitochondrial genome
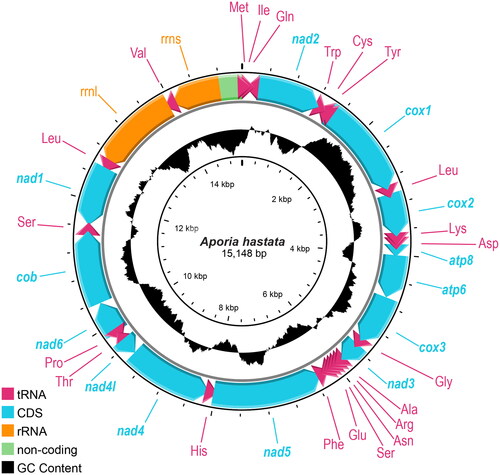
The mitochondrial genome of A. hastata is 15,148 bp in length (GenBank accession number: OP373108), with 40.23% A, 40.20% T, 7.29% G, and 12.28% C. The genome contains 13 PCGs, 22 tRNAs, and two rRNAs, plus a non-coding control region. The plus (+) strand encodes nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cob, while the minus (−) strand encodes nad5, nad4, nad4l, and nad1 (). The gene order of A. hastata is identical to other Aporia species.
3.2. Phylogenetic position
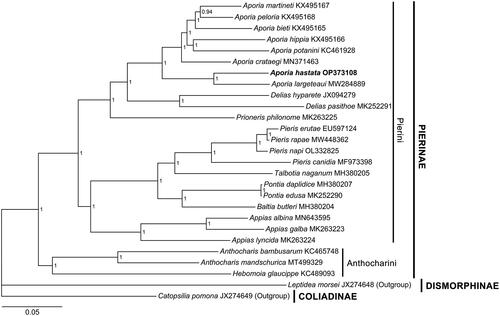
The phylogenetic results show that A. hastata is sister to A. largeteaui (Oberthür, 1881) and then clusters with other Aporia species in the A. crataegi group, namely A. crataegi (Linnaeus, 1758), A. potanini Alphéraky, 1892, A. hippia (Bremer, 1861), A. bieti (Oberthür, 1884), A. peloria (Hewitson, 1853) and A. martineti (Oberthür, 1884), with maximal node support values. All Aporia taxa are sisters to the genus Delias Hübner, [1819] within Pierini ().
4. Conclusion
The mitochondrial genome of A. hastata is 15,148 bp in length, with the gene order identical to other Aporia mitochondrial genomes (). The Bayesian Inference placed it with all available Aporia mitochondrial genomes in the tree (). Taxonomically, A. hastata and A. largeteaui are not sibling species, belonging to two different species groups (Della Bruna et al. Citation2013). However, due to a lack of mitogenomic data on other species in the group that A. hastata belongs to (the A. acraea group), the phylogenetic position of A. hastata is subject to change with more data in further research. Similarly, the topology of the cluster containing the A. crataegi group must also be revised when the mitogenome of A. genestieri (Oberthür, 1902) becomes available.
Ethical approval
The focal species used in this study is not under protection of CITES or wildlife laws in China, its status is not assessed by IUCN. The collecting and handling of the butterfly was conducted in accordance with the “Regulation on Experimental Animals of Yunnan Province.”
Authors’ contribution
S.-J. H. conceptualized and designed this study, Y.-Q. J. and S.-J. H. collected samples, Y.-Q. J. and X. Z. carried out experiment and data analysis, S.-J. H. and Y.-Q. J. drafted and revised the manuscript. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (44.1 KB)Supplemental Material
Download MS Word (324.1 KB)Acknowledgements
The authors thank Kuang Duan, Hui-Hong Zhang, and Dan Luo (Yunnan University, Kunming, China) for their assistance in field work and Adam M. Cotton (Chiang Mai, Thailand) for improving the earlier drafts of this article.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/genbank/, under accession no. OP373108. The associated Bio-Project and Bio-Sample numbers are PRJNA901130 and SAMN31704713 respectively. The present study used the Sanger dideoxy sequencing method and hence did not register the SRA in NCBI but provided the gel electrophoresis images (Figure S1) and Sanger sequencing results (*.ab1 files) in the Supplementary Package.
Additional information
Funding
References
- Back W. 2020. In: G. C. Bozano editor. Guide to the Butterflies of the Palearctic Region. Pieridae Part IV. Subfamily Pierinae (partim). Tribe Anthocharidini. Milano: Omnes Artes.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cao Y, Hao JS, Sun XY, Zheng B, Yang Q. 2016. Molecular phylogenetic and dating analysis of pierid butterfly species using complete mitochondrial genomes. Genet Mol Res. 15(4) gmr15049196.
- Della Bruna C, Gallo E, Sbordoni V. 2013. In: G. C. Bozano editor. Guide to the Butterflies of the Palearctic Region. Pieridae Part I. Subfamily Pierinae. Tribe Pierini (partim) 2 ed. . Milano: Omnes Artes.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kanoh K, Sakai S, Hayashi M. 2017. Radiative speciation in the genus Aporia (Pieridae, Lepidoptera). Evol Sci. 20:37–68.
- van Gasse P. 2021. Butterflies of the Indian Subcontinent - Distributional Checklist. Pardubice: Tshikolovets Publications.
- Wang WL, Suman DO, Zhang HH, Xu ZB, Ma FZ, Hu SJ. 2020. Butterfly conservation in China: from science to act. Insects. 11(10):661.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.