Abstract
Polygonatum infundiflorum Y.S. Kim, B.U. Oh & C.G. Jang et al. Citation1998 is a Korean endemic species. This is first report on the complete chloroplast genome sequence of P. infundiflorum. The complete chloroplast genome length was 154,578 bp with a GC content of 37.7%. The large single-copy (LSC) region was 83,527 bp long, and the small single-copy (SSC) region was 18,457 bp long. The paired inverted repeats (IRs) were 26,297 bp and separated the LCS and SSC regions. There were 113 genes, comprising 78 protein-coding genes, four rRNA genes, 30 tRNA genes, and one pseudogene (infA). In total, 16 genes contained one intron, and two genes contained two introns. Phylogenetic analysis suggested that Polygonatum was divided into three sections, each forming a monophyletic group. P. infundiflorum was sister to P. macropodum and formed a monophyletic group with P. inflatum. This study provides basic information for future research and contributes to taxonomic and genetic studies on Polygonatum.
Introduction
Polygonatum Mill. is belongs to Asparagaceae and is divided into three sections (Polygonatum, Verticillata, and Sibirica) based on phylogenetic studies (Meng et al. Citation2014; Floden and Schilling Citation2018). These species have been used in traditional medicine and are being actively studied in genetic systematics and marker development (Wujisguleng et al. Citation2012; Floden and Schilling Citation2018; Pan et al. Citation2020; Wang et al. Citation2022; Yang et al. Citation2022). Polygonatum infundiflorum Y.S. Kim, B.U. Oh & C.G. Jang et al. Citation1998 is a Korean endemic species discovered in Pungdo, Ansan-si, Gyeonggi-do (Jang et al. Citation1998; Oh et al. Citation2016). It belongs to the sect. Polygonatum, and can be distinguished from related species based on characteristics including curved stem tip, absence of trichomes along the midrib and margin on the abaxial leaf surface, erect stamens, and multicellular trichomes on filaments (Jang et al. Citation1998; Jang Citation2002; ). Although several studies have reported the genetics and complete chloroplast genomes of other Polygonatum species, few studies related to this species have been published. Therefore, we report the complete chloroplast genome of P. infundiflorum in this study to contribute to plant taxonomic identification, genetic diversity, and medicinal uses.
Materials and methods
P. infundiflorum was collected from Daejeon, Korea (39°19′20.8ʺN, 127°28′57.1ʺE), and voucher specimens were stored at Korea National Arboretum herbarium (http://www.nature.go.kr/kbi/plant/smpl/KBI_2001_030100.do; received by Hee Young Gil, E-mail: [email protected]) under the voucher number ESK21-311. Fresh leaves were dried using silica gel.
DNA was extracted using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) and DNA quality and concentration were confirmed through NanoDrop 2000 microspectrophotometer (Thermo Fisher Inc., Waltham, MA, USA). Next-generation sequencing (NGS) was generated for sequencing and data analysis using the TruSeq DNA PCR Free (550) library kit on the Illumina MiSeq platform (Macrogen, Inc., Seoul, Korea). The chloroplast sequence was assembled using GetOrganelle with ‘embplant_pt’ as a reference (Jin et al. Citation2020) and finally confirmed in Geneious Prime (Kearse et al. Citation2012). The coding sequence (CDS) and transfer RNA (tRNA) were annotated using GeSeq (Tillich et al. Citation2017) and tRNAscan-SE software (Lowe and Chan Citation2016). Genome maps, cis-splicing genes, and trans-splicing genes were generated using CPGview (Liu et al. Citation2023). The assembled sequence was registered in NCBI GenBank under the accession number OP764686.
We analyzed 30 Polygonatum species together with Maianthemum, Convallaria, and Dracaena of outgroups. We used the MAFFT program in PhyloSuite for 78 CDS sequences that were extracted, concatenated, and aligned (Zhang et al. Citation2020). We used the ModelFinder program in PhyloSuite (Kalyaanamoorthy et al. Citation2017) to find the best-fit models for the maximum likelihood (ML). The best-fit model of ML was TVM + F+R2 (AICc). The aligned sequence dataset was subjected to ML analysis using IQ-Tree with 10000 bootstrap replicates (Ronquist et al. Citation2012; Nguyen et al. Citation2015).
Results and discussion
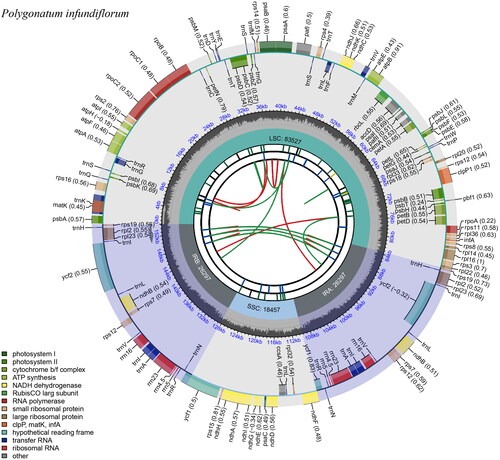
Of 7,914,172 reads, 33,856 reads were used to assemble the chloroplast genome of P. infundiflorum (maximum coverage: 115×, average coverage: 65.9×; Supplementary Figure 1). The sequence length of the complete chloroplast genome was 154,578 bp with a GC content of 37.7%. The length of the large single copy (LSC) region was 83,527 bp and that of the small single copy (SSC) region was 18,457 bp. The paired inverted repeats (IRs) were 26,297 bp, and separated the LCS and SSC regions (). A total of 113 genes existed, comprising 78 protein-coding genes, four rRNA genes, 30 tRNA genes, and one pseudogene (infA). The infA gene has a premature stop codon and has also been identified in Convallaria and Dracaena within the Asparagaceae family (Zhu et al. Citation2018; Raman et al. Citation2021). In total, 16 genes (10 protein-coding genes: atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, and rps16; six tRNA: trnA-UGC, trnG-UCC, trnl-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron and two genes (clpP1 and pafI) contained two introns. The rps12 is a trans-splicing gene (Supplementary Figure 2B) and thirteen genes including rps16, atpF, rpoC1, pafI, clpP1, petB, petD, rpl16, rpl2, ndhB, ndhA, ndhB, and rpl2 are cis-splicing genes (Supplementary Figure 2A).
Figure 2. Complete chloroplast circle map of Polygonatum infundiflorum drawn using CPGview (Liu et al. Citation2023). The circle map contains six tracks. The first track indicates the forward and reverse directions in red and green, respectively. The second track is a blue bar representing tandem repeats. The third track represents microsatellite sequences in green and yellow. LSC, SSC, and inverted repeat (IRa and IRb) regions are shown on the fourth track. The fifth track represents GC contents along plasma, and the sixth track represents genes.
The ML tree for the 78 CDS sequences were analyzed including 30 Polygonatum species and four outgroup species (Maianthemum, Convallaria, and Dracaena). The concatenated CDS length was 64476 bp. In the phylogenetic tree, outgroups branched separately, and Polygonatum was divided into three sections, each forming a monophyletic group. P. infundiflorum was sister to P. macropodum and formed a monophyletic group with P. inflatum. This clade indicated good support (BS = 100% and 88%, respectively), suggesting a close phylogenetic relationship between these three species ().
Figure 3. Maximum likelihood (ML) tree of 34 chloroplast genomes of Polygonatum and the outgroups. Values indicated above the branches are ML bootstrap support values (bootstrap repeat: 10000).
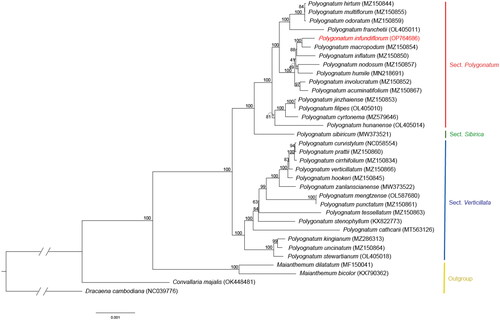
This study provides the cp genome information of P. infundiflorum, which would contribute to the taxonomic and genetic studies on Polygonatum.
Ethical approval
This study complied with relevant institutional, national, and international guidelines and legislation. This species is not an endangered or protected species, and the collection area is not protected.
Author contributions
Se Ryeong Lee analyzed the data and drafted the manuscript. Sang-Chul Kim collected and analyzed the data. Young-Ho Ha collected the data and conceived the original structure of the review along with Dong Chan Son. All the authors have read and agreed to the submitted version of the manuscript.
Supplemental Material
Download JPEG Image (1.8 MB)Supplemental Material
Download JPEG Image (209 KB)Supplemental Material
Download MS Word (486.6 KB)Acknowledgments
We thank Sa-Bum Jang and Eun-Ho Lee for their assistance in sampling and laboratory work throughout the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome data that support the findings of this study are available at NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OP764686. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA897089, SRR22184768, and SAMN31577749, respectively.
Additional information
Funding
References
- Floden A, Schilling EE. 2018. Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol Phylogenet Evol. 129:202–213.
- Jang CG, Oh BU, Kim YS. 1998. A new species of Polygonatum from Korea; P. grandicaule. Korean J Pl Taxon. 28(1):41–47.
- Jang CG. 2002. A taxonomic review of Korean Polygonatum (Ruscaceae). Korean J Pl Taxon. 32(4):417–447.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 23(3):694–704.
- Lowe TM, Chan PP. 2016. TRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Meng Y, Nie ZL, Deng T, Wen J, Yang YP. 2014. Phylogenetics and evolution of phyllotaxy in the Solomon’s seal genus Polygonatum (Asparagaceae: polygonateae). Bot J Linn Soc. 176(4):435–451.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oh BU, Ko SC, Kang SH, Paik WK, Yoo KO, Im HT, Jang CG, Chung GY, Choi BH, Choi HJ, et al. 2016. Distribution Maps of Vascular Plants in Korea. Pocheon:Korea National Arboretum,. pp. 1–809.
- Pan J, Lu W, Chen S, Cao T, Chi L, He F. 2020. Characterization of the complete chloroplast genome of Polygonatum sibiricum (Liliaceae), a well-known herb to China. Mitochondrial DNA Part B Resour. 5(1):528–529.
- Raman G, Lee EM, Park SJ. 2021. Intracellular DNA transfer events restricted to the genus Convallaria within the Asparagaceae family: possible mechanisms and potential as genetic markers for biographical studies. Genomic. 113(5):2906–2918.
- Ronquist F, Teslenko M, van der Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, Qian J, Jiang Y, Chen X, Zheng B, Chen S, Yang F, Xu Z, Duan B. 2022. Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of Polygonatum and tribe Polygonateae. Front Plant Sci. 13:882189.
- Wujisguleng W, Liu Y, Long CH. 2012. Ethnobotanical review of food uses of Polygonatum (Convallariaceae) in China. Acta Soc Bot Pol. 81(4):239–244.
- Yang Q, Jiang Y, Wang Y, Han R, Liang Z, He Q, Jia Q. 2022. SSR Loci analysis in transcriptome and molecular marker development in Polygonatum sibiricum. Biomed Res Int. 2022ID4237913::4237913.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhu Z-X, Mu W-X, Wang J-H, Zhang J-R, Zhao K-K, Friedman CR, Wang H-F. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondrial DNA Part B Resour. 3(2):620–621.

