Abstract
Lemmaphyllum carnosum var. drymoglossoides (Baker) X. P. Wei, 2013 is a valuable medicinal fern in China. Its complete chloroplast genome was determined using Illumina paired-end sequencing. The genome was 157,571 bp in length with 130 genes, including 87 protein-coding genes, eight ribosomal RNA genes, and 35 tRNA genes. It displayed a quadripartite structure consisting of a small single-copy (SSC) of 21,691 bp, a large single-copy (LSC) of 81,106 bp, and two inverted repeats (IRs) of 27,387 bp, respectively. The phylogenetic results indicated that L. carnosum var. drymoglossoides exhibited the closest relationship with L. intermedium, and this study provided new information for the phylogenetic relationship of the Polypodiaceae family.
Introduction
Lemmaphyllum carnosum var. drymoglossoides (Baker) X. P. Wei, 2013, an epiphytic fern with medicinal value belonging to the Polypodiaceae family (Zhang et al. Citation2014), was first reported in 1940 with the name Lepidogrammitis drymoglossoides (Baker) Ching 1940 (Wei and Zhang Citation2013) (). This species is epiphytic on shaded tree trunks and rocks, 200–1400 m above altitude, and is widely distributed in all provinces south of the Yangtze River basin. The entire plant is used as a traditional herbal medicine in Southwest China and has important applications in ethnomedicine (Chen et al. Citation2000). It is effective in clearing away heat and toxins, relieving dampness, and eliminating stasis, and it is also used for treating bronchitis, pleurisy, tuberculosis, dysentery, and rheumatoid arthritis (Lin et al. Citation2000, Yan and Yu Citation1998). L. carnosum var. drymoglossoides is easily confused with the L. carnosum var. microphyllum, and they can only be distinguished by the discrete sori in contrast to the linear coenosori of L. carnosum var. microphyllum (Lin et al. Citation2000). Chloroplast genomes have been widely used in species delimitation and phylogeny because of their uniparental inheritance and lower substitution rates than nuclear genomes (Wei et al. Citation2020, Gu et al. Citation2022). This study characterizes the first complete chloroplast genome of L. carnosum var. drymoglossoides using high throughput sequencing technology and reconstructs the phylogenetic relationships utilizing the published chloroplast genome sequences of the Polypodiaceae family.
Figure 1. Plant image of L. carnosum var. drymoglossoides. Sterile leaves oblong or ovate, rounded or obtusely rounded, base cuneate, entire. Fertile leaves ligulate or oblanceolate, narrowly constricted at the base, sometimes the same shape as the sterile leaves, fleshy, leathery when dry. This photograph was taken by Chong-Jian Zhou in Huangshan City, Anhui Province, and was used with the author’s permission.

Materials and methods
L. carnosum var. drymoglossoides was collected from Fuxi Village (Tangkou Town, Huangshan City, Anhui Province, China N30°07′, E118°12′) and deposited at Chengde Medical University (Jinxin Liu, [email protected]) with the voucher number HPAA0005. The total genomic DNA was isolated from fresh, healthy fronds using modified CTAB methods. The DNA quality was estimated using a Qubit 4.0 Fluorometer (Thermo Fisher Scientific Inc., USA), followed by shearing to prepare a PCR-free library of 350 bp. High-throughput sequencing was performed using the Illumina NovaSeq 6000 system, generating a total of 2.2 G of pair-end raw data. Trimmomatic v0.38 (Bolger et al. Citation2014) was used to clean the sequencing adapters and low-quality reads. The clean data was then de novo assembled with GetOrganelle v1.7.3.5 (Jin et al. Citation2020). The depth of coverage was calculated by mapping the reads onto the chloroplast genome sequence with bowtie2 v2.3.4.3 to determine the correctness of the assembly (Langmead and Salzberg Citation2012). The assembled chloroplast genome was annotated using the GeSeq (Tillich et al. Citation2017) and CPGAVAS2 online webserver (www.herbal genomics.org/cpgavas2) (Shi et al. Citation2019), and corrected via a blasting search against Lemmaphyllum intermedium and L. carnosum var. microphyllum (GenBank: NC_053788, MN623356) as references. Chloroplast Genome Viewer (CPGView) was used to draw the circular map of chloroplast genome (Liu et al. Citation2023).
Result
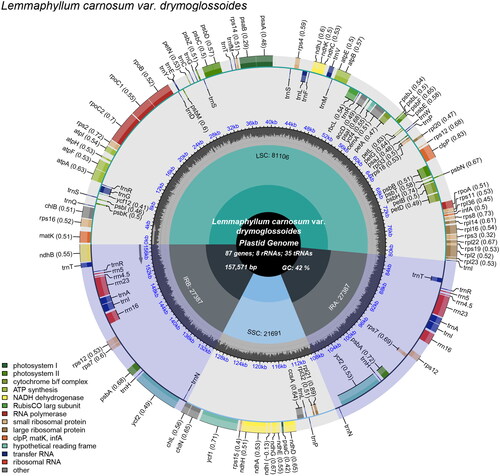
The chloroplast genome of L. carnosum var. drymoglossoides is 157,571 bp in length, with an average depth of 513.14 X (Supplementary Figure 1). The depth of some ares are about 80X because the low-complexity regions affect the efficiency of reads mapping, such as ‘AATACCCCCCCCCC’. The chloroplast genome of L. carnosum var. drymoglossoides was circular with a quadripartite structure consisting of a small single-copy (SSC) of 21,691 bp, a large single-copy (LSC) of 81,106 bp, and two inverted repeats (IRs) of 27,387 bp, respectively (). Furthermore, the L. carnosum var. drymoglossoides chloroplast genome encoded 130 genes, including 87 protein-coding genes, eight rRNA genes, and 35 tRNA genes. Gene structure analysis was done for rps16, atpF, rpoC1, clpP, petB, petD, rpl16, rpl2, ndhA, ndhB difficult to annotate genes (Supplementary Figure 2). The GC content of the entire genome was 41.8%, displaying LSC, SSC, and IR GC levels of 40.5%, 37.9%, and 45.3%, respectively. One intron occurred in 16 genes: rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, clpP, petB, petD, rpl16, ndhB, trnT-UGU, trnA-UGC, trnI-GAU, ndhA, trnI-GAU, and rpl2. Only the clpP gene displayed two intors. Fifteen genes were duplicated in the IR regions, namely ndhB, trnT-UGU, trnR-ACG, rrn5S, rrn4.5S, rrn23S, trnA-UGC, trnI-GAU, rrn16s, rps12, rps7, psbA, trnH-GUG, ycf2, and trnN-GUU.
Figure 2. Chloroplast genome map of L. carnosum var. drymoglossoides. Genes drawn outside the outer circle are transcribed counterclockwise, and genes drawn inside the outer circle are transcribed clockwise. Genes belonging to diferent functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions.
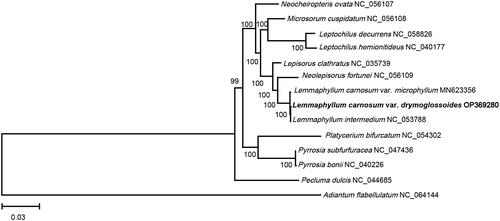
To conduct the phylogenetic analyses, the complete chloroplast genome sequences of 13 ferns were retrieved from the National Center for Biotechnology Information (NCBI). The alignment of 80 protein-coding genes was first created using the muscle v5 (Edgar Citation2022). and then concatenated to a super alignment with a length of 67900 bp. A maximum likelihood (ML) tree was constructed with RAxML v8.2.12 (Stamatakis Citation2014) using Adiantum flabellulatum as an outgroup. Furthermore, 14 chloroplast genomes of 14 species, including 13 species in the Polypodiaceae family, and one species (Adiantum flabellulatum) in the Adiantaceae family in the Cornaceae family, were used for phylogenetic analysis. The results indicated that L. carnosum var. drymoglossoides exhibited the closest relationship with L. intermedium with a high bootstrap value of 100 (). This study provides new evidence for the phylogenetic study of the genus Lemmaphyllum.
Figure 3. The phylogenetic position for L. carnosum var. drymoglossoides according to the ML phylogenetic tree constructed based on 14 chloroplast genomes. The following sequences were used: Lemmaphyllum carnosum var. drymoglossoides OP369280, Adiantum flabellulatum NC_064144, Lemmaphyllum carnosum var. microphyllum MN623356 (Liu et al. Citation2020), Lepisorus clathratus NC_035739 (Wei et al. Citation2017), Leptochilus decurrens NC_058826 (Su et al. Citation2019), Leptochilus hemionitideus NC_040177 (Min et al. Citation2018), Lemmaphyllum intermedium NC_053788 (Wang et al. Citation2021), Microsorum cuspidatum NC_056108, Neolepisorus fortunei NC_056109, Neocheiropteris ovata NC_056107 (Liu et al. Citation2021), Pecluma dulcis NC_044685 (Samuli and Cárdenas Citation2019), Platycerium bifurcatum NC_054302, Pyrrosia bonii NC_040226 (Cai et al. Citation2018), Pyrrosia subfurfuracea NC_047436 (Min et al. Citation2019). The sequences used for the tree structure are coding sequences. The bootstrap support values are shown on the nodes.
Discussion and conclusion
In this study, the chloroplast genome sequence of L. carnosum var. drymoglossoides was assembled for the first time and the structure of this species was annotated. The phylogenetic results indicated that L. carnosum var. drymoglossoides exhibited the closest relationship with L. intermedium, and this study provided new information for the phylogenetic relationship of the Polypodiaceae family.
Ethical approval
The material involved in the article does not involve ethical conflicts. This species is neither endangered on the cites catalogue nor collected from a natural reserve, so it did not need specific permissions or licenses. All collection and sequencing work was strictly executed under local legislation and related laboratory regulations to protect wild resources.
Author contributions
JXL and LCS conceived and designed the experiments; HYZ and XYL performed the experiments; ZLZ and YT analyzed the data and modified the article; ZLZ and HYZ wrote the paper. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (273.3 KB)Supplemental Material
Download TIFF Image (150.5 KB)Supplemental Material
Download MS Word (376.3 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genom esequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP369280. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682118, SRR23238769, and SAMN32925244, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cai S, Cai X, Li S, Liu S, Wang Z, Wang T, Su Y. 2018. The complete chloroplast genome of Pyrrosia bonii (Polypodiaceae), an important ornamental and medical fern. Mitochondrial DNA B Resour. 3(2):801–802.
- Chen Z, Li C, Wang M. 2000. Antiinflammation By Water Extraction of Lepidogrammitis Drymoglossoides (bak.) Ching. Strait Pharm J.: 37–38.
- Edgar RC. 2022. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun. 13:6968.
- Gu X, Hao D, Xiao P. 2022. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin Herb Med. 14(2):171–186.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Lin Y, Zhang X, Shi L, Lu S. 2000. Flora of China. Beijing: Science Press. 6(2):96–97.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu S, Wang Z, Su Y, Wang T. 2021. Comparative genomic analysis of Polypodiaceae chloroplasts reveals fine structural features and dynamic insertion sequences. BMC Plant Biol. 21(1):31.
- Liu S, Wang Z, Wang H, Su Y, Wang T. 2020. Patterns and rates of plastid rps12 gene evolution inferred in a phylogenetic context using plastomic data of ferns. Sci Rep. 10(1):9394.
- Min Y, Cai S, Xiao H, Zhang M, Hong Y, He Z, Wang Z, Wang T, Su Y. 2019. The complete chloroplast genome of Pyrrosia calvata (Polypodiaceae), a traditional Chinese medicinal fern only restricted to Guangxi, China. Mitochondrial DNA Part B. 4(1):1757–1758. J. M. D. P. B.
- Min Y, Guan J, Li S, Liu S, Hong Y, Wang Z, Wang T, Su Y. 2018. The complete chloroplast genome of Leptochilus hemionitideus, a traditional Chinese medical fern. Mitochondrial DNA B Resour. 3(2):784–785.
- Samuli L, Cárdenas G. 2019. Dynamism in plastome structure observed across the phylogenetic tree of ferns. Botan J Linnean Soc. 190(3):229–241.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–w73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su Y, He Z, Wang Z, Hong Y, Wang T. 2019. Characterization of the complete chloroplast genome of Leptochilus decurrens (Polypodiaceae), a least concern folk medicinal fern. Mitochondrial DNA B Resour. 4(2):3346–3347.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–w11.
- Wang Z, Li N, Li Z, Liu Y, Yang P, Hong Y, He Z, Su Y, Wang T. 2021. The complete chloroplast genome of Lemmaphyllum intermedium, a valuable medicinal fern. Mitochondrial DNA B Resour. 6(2):423–424.
- Wei R, Yan YH, Harris AJ, Kang JS, Shen H, Xiang QP, Zhang XC. 2017. Plastid Phylogenomics Resolve Deep Relationships among Eupolypod II Ferns with Rapid Radiation and Rate Heterogeneity. Genome Biol Evol. 9(6):1646–1657.
- Wei XP, Zhang XC. 2013. Species delimitation in the fern genus Lemmaphyllum (Polypodiaceae) based on multivariate analysis of morphological variation. Jnl of Sytematics Evolution. 51(4):485–496.
- Wei XP, Li HJ, Che P, Guo HJ, Zhang BG, Liu HT, Qi YD. 2020. Comparing chloroplast genomes of traditional Chinese herbs Schisandra sphenanthera and S. chinensis. Chin Herb Med. 12(3):247–256.
- Yan YQ, Yu CL. 1998. Encyclopedia of Chinese Medicine, Beijing: China Medical Science and Technology Press.
- Zhang L, Ren L, Wang T, Dong X, Wan M, Wu H, Mei Q, Gao Y. 2014. Chemical constituents from Lepidogrammitis drymoglossoides. Chinese Herbal Medicines. 45:2890–2894.
