Abstract
Phyllostachys incarnata Wen, 1982 is one of the important material and edible bamboo specie of high quality in China. We reported the complete chloroplast(cp) genome of P. incarnata in this study. The cp genome of P. incarnata (GenBank accession number: OL457160) was a typical tetrad structure with a full length of 139,689 bp, comprising a pair of inverted repeated (IR) regions (21,798 bp) separated by a large single-copy (LSC) region (83,221 bp) and a small single-copy (SSC) region (12,872 bp). And the cp genome contained 136 genes, including 90 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on 19 cp genomes suggested that P. incarnata was relatively close to P. glauca among the species analyzed.
Introduction
Phyllostachys incarnata Wen, 1982 is a bamboo species with high-quality shoots, which belongs to Phyllostachys of Poaceae Barnhart and originated from Fujian and Zhejiang provinces of China (Chen et al. Citation2006). Currently, it is also distributed in Sichuan, Anhui, Jiangxi and other regions for large scale introduction and cultivation with a high priority, because of its high yield and long growing season of the edible shoots. It has been praised for characteristics of early emergence (April–May), strong shooting ability, long duration and excellent shoot quality. Furthermore, this bamboo species provides pest and disease resistance, and tolerance of abiotic stresses such as drought, cold, and waterlogging. And beyond that, it can also allow gardeners to support cultivated landscapes with unique nature (Leng Citation2017).
Typical characteristics of P. incarnata including the young bamboo poles were thickly white powdery especially below nodes; sheaths abaxially fleshy red or greenish or distally green on slender culms, with sparse speckles; final branchlets with 3 or 4 leaves; auricles flourishing, ovate or semicircular, greenish purplish, with radial tassels; ligule strongly projecting, purplish, acuminate upward (). Chloroplast genes are related to many important traits in plants, including resistance to herbicides, insect resistance, and stress tolerance (Daniell et al. Citation2005). Currently, chloroplast gene engineering has been applied to improve plant resistance to herbicides and insects, as well as to increase their stress tolerance (Wani et al. Citation2015). Hence, analyzing the chloroplast genes of P. incarnata will aid in comprehending its adaptability and expedite the enhancement of its varieties. In this study, we report and characterize the chloroplast genome of P. incarnata. Using these data, we reconstruct the phylogenetic tree of this species to reveal the relationship and provide useful information for further study of P. incarnata.
Materials and methods
Plantlet of P. incarnata were collected from Wenjiang District, Chengdu City, Sichuan Province, China (N30°42′6″, E103°51′30″) on 7 September 2021. The specimens were deposited in the herbarium room of Aba Teachers University, Aba, Sichuan, China, China (http://www.abtu.edu.cn/; contact person: LiHua Wang; Email: [email protected]) under the voucher number ATUP02109070002. Total DNA was extracted from the fresh leaves of P. incarnata using NEBNext Ultra DNA Library Prep Kit for Illumina (E7370L, New England Biolabs), and the high-quality DNA was sheared to the fragments of 350 bp in length for the shotgun library construction, which were sequenced using the Illumina NovaSeq platform (Illumina Inc, San Diego, CA), and thus generating 150 bp paired-end reads. The filtered reads were assembled into the complete chloroplast genome using the program A5-miseq v20150522 (Coil et al. Citation2015) and SPAdes v3.9.0 (Bankevich et al. Citation2012) and with P. edulis chloroplast genome (GenBank accession number: NC_015817) as a reference. The annotation of chloroplast genome was conducted through the online program CPGAVAS2. The whole chloroplast genome map was drawn using CPGView (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023). The annotated genomic sequence has been registered into GenBank with the accession number (OL457160).
Results
The cp genome of P. incarnata was a typical tetrad structure with a full length of 139,689 bp, comprising a pair of inverted repeated (IR) regions (21,798 bp) separated by a large single-copy (LSC) region (83,221 bp) and a small single-copy (SSC) region (12,872 bp) (). A total of 136 genes, including 90 protein-coding genes, 38 tRNA genes and 8 rRNA genes, are successfully annotated in the complete chloroplast genome sequence of P. incarnata. And the complete chloroplast genome contains 109 unique genes, including 80 protein-coding genes, 29 tRNA genes and 4 rRNA genes. The rps12 is a trans-spliced gene (Figure S1). It has three unique exons. Two of them are duplicated as they are located in the IR regions. Eleven genes, including rps16, atpF, ycf3, petB, petD, rpl16, rpl12, ndhB, ndhA, ndhB, and rpl2, contain one or two introns (Figure S2). The complete genome GC content was 38.89% (Table S1), while the corresponding values of the LSC, SSC and IR were 36.97%, 33.16% and 44.23%, respectively.
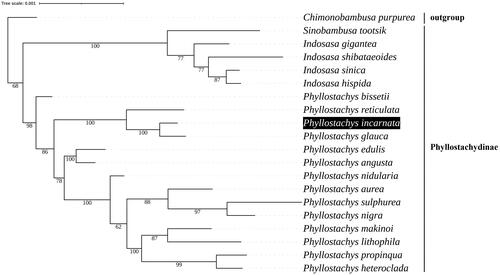
The maximum-likelihood phylogenetic tree was constructed based on 19 complete chloroplast genomes of Phyllostachydinae species, and Chimonobambusa purpurea as outgroup. All sequences were obtained from NCBI GenBank. The complete protome nucleotide sequences were extracted, aligned and concatenated in PhyloSuite v1.2.2 (Zhang et al. Citation2020), and used to perform the ML inference in IQ-TREE Multicore version 1.6.12 (Gao et al. Citation2018). Phylogenetic analysis suggested that P. incarnata was relatively close to P. glauca among the species analyzed ().
Figure 2. Schematic map of overall features of P. incarnata chloroplast genome. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The base frequency at each site along the genome will be shown between the fourth and fifth tracks. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively.
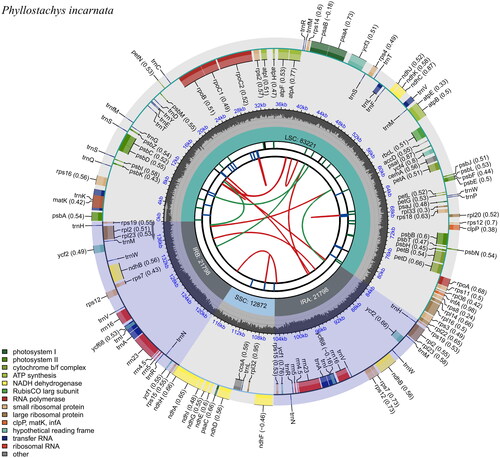
Figure 3. Maximum-likelihood phylogenetic analysis of 19 species of Subtrib. Phyllostachydinae and Chimonobambusa purpurea as outgroup were based on complete protome nucleotide sequences. Bootstrap values were indicated below the nodes. The following sequences were used for tree construction: P. glauca (NC_051535.1) (Cao et al. Citation2020), P. reticulata (MN537808.1) (Huang et al. Citation2019), P. sulphurea (NC_024669.1) (Gao and Gao Citation2016), P. edulis (NC_015817.1) (Huang et al. Citation2019), P. nidularia (LC590826.1) (Zhou et al. Citation2021), P. nigra (NC_015826.1) (Zheng et al. Citation2021), P. heteroclada (NC_064526.1) (Hu et al. Citation2021), P. angusta (NC_053647.1) (Zheng et al. Citation2021), P. propinqua (NC_016699.1) (Wu and Ge Citation2012), P. makinoi (NC_062168.1), P. lithophila (NC_062169.1), P. bissetii (NC_066717.1), P. aurea (KU569973.1) (Attigala et al. Citation2016), P. incarnata (OL457160), Indosasa gigantea (NC_046587.1) (Zheng et al. Citation2020), I. sinica (NC_024721.1) (Ma et al. Citation2014), I. hispida (MW463058.1) (Tu et al. Citation2022), I. shibataeoides (NC_036820.1) (Ma et al. Citation2017), Sinobambusa tootsik (MN783350.1), Chimonobambusa purpurea (NC_060376.1) (Liu et al. Citation2021).
Discussion
The chloroplast genome of plants has a very conserved structure and genetic composition (Maier et al. Citation1995; Daniell et al. Citation2016; Yan et al. Citation2023). In this study, we obtained and analyzed the chloroplast genome of P. incarnata for the first time. We identified that the genomic structures, gene contents and orders were highly conserved and similar to other Phyllostachydinae species (Attigala et al. Citation2016, Hu et al. Citation2021, Huang et al. Citation2019, Zhou et al. Citation2021, Zheng et al. Citation2021, Zheng et al. Citation2021, Wu and Ge Citation2012, Zheng et al. Citation2020, Ma et al. Citation2014, Tu et al. Citation2022, Ma et al. Citation2017, Liu et al. Citation2021). We also analyzed the phylogenetic relationships of P. incarnata by complete protome nucleotide sequences, which can provide valuable insights into the phylogenetic and evolutionary position of P. incarnata in the Phyllostachydinae subtribes and Gramineae family.
Ethical approval
The study was approved by the institutional review board of Aba Teachers University, Aba, Sichuan, China. The collection of plant materials was conducted in accordance with guidelines provided by the Aba Teachers University and Sichuan province regulations. Field studies complied with Sichuan province legislation.
Author contributions
H. Wang, W. Liu, B.X. Wang, Y.K. Liu and J.N. Wang designed research study and obtained the funding. L.H. Wang and W. Liu carried out the experiment. B.X. Wang wrote the manuscript with support from Y.K. Liu offered great in data analysis and all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (965 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OL457160. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA893462, SRR22044020, and SAMN31424863 respectively.
Additional information
Funding
References
- Attigala L, Wysocki WP, Triplett JK, Duvall MR, Clark LG. 2016. Phylogenetic estimation and morphological evolution of Arundinarieae (Bambusoideae: poaceae) based on plastome phylogenomic analysis. Mol Phylogenet Evol. 101:111–121.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cao BB, Tingting Ge TT, Shixiong Ding SX, Guo CC. 2020. The complete chloroplast genome of Phyllostachys glauca (Bambusoideae), a dominant bamboo species in limestone mountains endemic to China. Mitochondrial DNA B Resour. 5(3):3193–3194.
- Chen SL, Li DZ, Zhu GH, Wu ZL, Lu SL, Liu L, Wang ZP, Sun BX, Zhu ZD, Xia NH, et al. 2006. Flora of China 22. Beijing: Science Press. p. 174.
- Coil D, Jospin G, Darling AE. 2015. A5-MiSeq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Daniell H, Kumar S, Dufourmantel N. 2005. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 23(5):238–245.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi:10.1186/s13059-016-1004-2.
- Gao J, Gao LZ. 2016. The complete chloroplast genome sequence of the Phyllostachys sulphurea (Poaceae: bambusoideae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):983–985.
- Gao FL, Shen JG, Liao FR, Cai W, Lin SP, Yang HK, Chen SL. 2018. The first complete genome sequence of narcissus latent virus from Narcissus. Arch Virol. 163(5):1383–1386.
- Hu YP, Zhou J, Yu ZY, Li JJ, Xu MY, Guo QR. 2021. The complete chloroplast genome of Phyllostachys heteroclada f. solida (Poaceae). Mitochondr DNA B Resour. 6(2):566–567.
- Huang NJ, Li JP, Yang GY, Yu F. 2019. Two plastomes of Phyllostachys and reconstruction of phylogenic relationship amongst selected Phyllostachys species using genome skimming. Mitochondrial DNA B Resour. 5(1):69–70.
- Leng HB. 2017. The comprehensive evaluation of introduction value of 20 ornamental bamboo species. J Bamb Res. 36(2):21–28.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu W, Zhang J, Liu YK, Huang QY, Xiao QG, Wang BX. 2021. The complete chloroplast genome of Chimonobambusa purpurea (Bambusatae). Mitochondrial DNA B Resour. 6(2):691–6 92.
- Ma PF, Vorontsova MS, Nanjarisoa OP, Razanatsoa J, Guo ZH, Haevermans T, Li DZ. 2017. Negative correlation between rates of molecular evolution and flowering cycles in temperate woody bamboos revealed by plastid phylogenomics. BMC Plant Biol. 17(1):260.
- Ma PF, Zhang YX, Zeng CX, Guo ZH, Li DZ. 2014. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol. 63(6):933–950.
- Tu DD, Hui CM, Zhu LY, Zhang WJ, Liu WY. 2022. The complete chloroplast genome of Indosasa hispida ‘Rainbow’ (Poaceae, Bambuseae): an ornamental bamboo species in horticulture. Mitochondrial DNA B Resour. 7(4):619–621.
- Maier RM, Neckermann K, Igloi GL, Kössel H. 1995. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol.251(5):614–28.
- Wani SH, Sah SK, Sági L, Solymosi K. 2015. Transplastomic plants for innovations in agriculture. Agron Sustain Dev. 35(4):1391–1430.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62(1):573–578.
- Yan L, Yang XL, Wu YF, Wang X, Hu XJ. 2023. Chloroplast genomic characterization and phylogenetic analysis of Pellionia scabra. Chin J Biotech.
- Zhang D, Gao FL, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zheng X, Yang M, Ding YL, Lin SY. 2020. The complete chloroplast genome sequence of Acidosasa gigantea (Bambusoideae: Arundinarieae): an ornamental bamboo species endemic to China. Mitochondrial DNA B Resour. 5(1):1119–1121.
- Zheng Y, Zhao LY, Wang T, Chen GF, Hong YF, Yue JJ. 2021. The complete chloroplast genome of Phyllostachys angusta McClure (Poaceae). Mitochondrial DNA B Resour. 6(1):187–188.
- Zhou J, Hu YP, Yu ZY, Li JJ, Xu MY, Guo QR. 2021. The complete chloroplast genome of a solid type of Phyllostachys nidularia (Bambusoideae: poaceae), a species endemic to China. Mitochondr DNA B Resour. 6(3):978–979.

