Abstract
The perennial herbal medicine species Aconitum tschangbaischanense, is endemic to Changhai Mountain, Jilin province. In this study, we attempted to uncover the complete chloroplast (cp) genome of A. tschangbaischanense based on sequencing data using the Illumina sequencing technology. As per the results: (1) the length of its complete cp genome is 155,881 bp with a typical tetrad structure; (2) the structure of its cp genome contains large single-copy and small single-copy (LSC and SSC) regions of 86,351 and 16,9444 bp, respectively, isolated by two inverted repeat regions (IRs) of 26,293 bp; (3) we annotated a total 131 genes, consisting of 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. According to the maximum-likelihood phylogenetic tree based on complete cp genomes, A. tschangbaischanense, showed close association with A. carmichaelii, which belongs to clade I. Finally, this study provides the characteristics of the cp genome of A. tschangbaischanense, and its phylogenetic position.
Aconitum (Ranunculaceae) is a perennial genus that includes more than 400 species and is mainly distributed in the temperate regions of the Northern Hemisphere (Li and Kadota Citation2001). China is one of the diversity centers for Aconitum with more than 200 species, and approximately 76 species are utilized in traditional Chinese medicine (Li and Kadota Citation2001; Xiao et al. Citation2006). Aconitum tschangbaischanense S. H. Li & Y. H. Huang, 1975, is an endemic species limited to the Changbai Mountain area between 1700 and 2100 m (Zhou Citation2009) (). It is only concentrated in the meadows of the Betula ermanii belt and alpine tundra belt in the Changbai Mountain National Nature Reserve. Species with narrow distributions are at risk of extinction. It is listed as an endangered species and is in urgent need of protection (Zhou Citation2006); however, it has not been assessed by the International Union for Conservation of Nature (IUCN). Therefore, it is necessary to study its genetic diversity to guide its ex situ and in situ conservation. We assembled the complete plastome of A. tschangbaischanense to (1) characterize and compare the complete plastome of A. tschangbaischanense and (2) assess the taxonomic position of A. tschangbaischanense.
We collected samples from the south slope of Changbai Mountain in Jilin province, China (128°2′E, 41°56′N, Alt.: 1850 m). This work was approved by the Changbai Mountain Nature Conservation Management Center and complies with the IUCN policy research involving species at risk of extinction (see Guidelines for appropriate uses of IUCN Red List Data), the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. The silica gel dried leaves and specimens were deposited at the herbarium of Tonghua Normal University (Xue-Lian Liu, [email protected]) under the voucher number LXL202115 (plant identification was completed by Professor You Zhou of Tonghua Normal University). Total genomic DNA was extracted from the dried leaves using the CTAB method (Doyle and Doyle Citation1987). Paired-end sequencing of whole sequences from both ends of 150 bp fragments was performed on the DNBSEQ T7 at Benagen (https://www.benagen.com), and approximately 4 Gb clean data were generated for A. tschangbaischanense. A total of 28,270,584 clean reads and 4,228,588,412 clean bases were produced for de novo assembly using the GetOrganelle pipeline (Jin et al. Citation2020). Aconitum kusnezoffii was used as a reference for annotation using the program CPGAVAS2 (Shi et al. Citation2019). We also used CPGview (http://www.1kmpg.cn/cpgview) to improve annotation (Liu et al. Citation2023). To identify the phylogenetic position of A. tschangbaischanense, the maximum-likelihood (ML) tree was reconstructed based on 40 species of complete chloroplast (cp) genomes using IQtree V2.1.3.
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under accession no. OP221050. The read coverage depth map is shown in Figure S1. The complete cp genome of A. tschangbaischanense has a quadripartite structure comprising 155,881 bp in length, with 38% of the overall guanine and cytosine (GC) content. There is a LSC region of 86,351 bp and an SSC region of 16,944 bp, which are separated by a pair of inverted repeat (IR) regions (IRa and IRb) of 26,293 bp in length. The GC contents of the corresponding values in the LSC, SSC, and IR regions were found to be 38.14%, 31.64%, and 45.23%, respectively ().
Figure 2. The map of A. tschangbaischanense chloroplast (cp) genome by CPGview. The map contains the core area and two tracks. The basic information of the cp genome shows in the core area. The GC content along the genome is plotted on the inner track. The genes showed on the external track, and genes with different functional groups are identified by different colors.
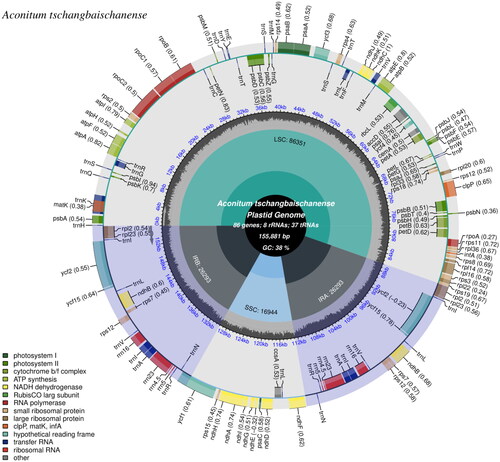
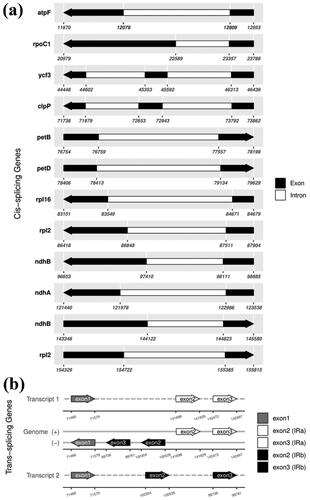
A total of 131 predicted genes in the cp genome of A. tschangbaischanense were assigned to three groups based on their functions: 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes (). Twelve cis-splicing gene were identified. Compared to Arabidopsis thaliana, the rps16 gene was not generated in the cp genome of A. tschangbaischanense (Liu et al. Citation2023; ). The structure of trans-splicing gene rps12 of A. tschangbaischanense showed there are three unique exons. Two of them are duplicated as they are located in the IR regions (). We also identified 35 simple sequence repeats, 32 long tandem repeats, and 19 long tandem repeats. The number of mono-, di-, tri-, and tetra-nucleotides SSRs was 22, 11, 1, and 1, respectively.
Figure 3. Schematic of the cis-splicing gene map (a) and trans-splicing genes map (b) generated for the chloroplast genome of A. tschangbaischanense. (a) The genes are arranged from left to right based on their order on the chloroplast genome. The gene names are shown on the top, and the gene structures are on the bottom. The exons are shown in black; the introns are shown in white. The arrow indicates the sense direction of the gene. Please note that lengths of exons and introns are not drawn to scale. (b) The start and end positions on the pre-mRNA are shown below the line. The lines represent the genome plus (+) and minus (−) DNA strands. The arrowheads represent the corresponding exons of the rps12 genes.
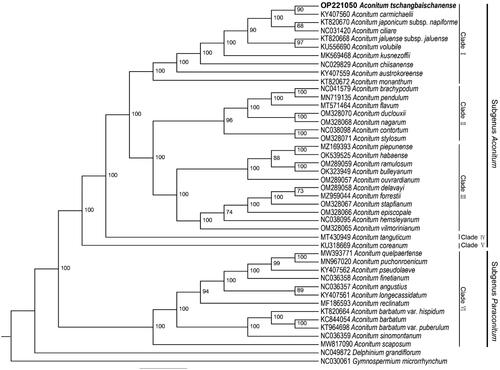
To understand the phylogenetic relationship of A. tschangbaischanense with related taxa in Aconitum, a ML tree was constructed using IQtree V2.1.3, based on the complete cp genomes of 40 Aconitum species and two outgroup species (Gymnospermium microrrhynchum and Delphinium grandiflorum) (Chen et al. Citation2015; Choi et al. Citation2016; Duan et al. Citation2019; Kim et al. Citation2022; Kong et al. Citation2017; Kong et al. Citation2018; Li et al. Citation2020; Lim et al. Citation2017; Lim et al. Citation2020; Liu et al. Citation2020; Ni et al. Citation2022; Park et al. Citation2017; Wang and Li Citation2020; Xia et al. Citation2022; Yang et al. Citation2022; Zhang et al. Citation2021). The ML tree showed that all the species in our study were divided into six main clades, belonging to two subgenera (subgenus Aconitum and Paraconitum). There are two clades with only one species, A. tanguticum and A. coreanum. Ten, seven, and 11 species were grouped into clades I, II, and III, respectively, which belonged to the subgenus Aconitum. Clade VI has 12 species and formed the subgenus Paraconitum. The plastid phylogenomic tree indicated that A. tschangbaischanense was closely related to A. carmichaelii, which belongs to the subgenus Aconitum ().
Author contributions
Zhao HY and Zhu JY were involved in the conception and design. Liu XL, Jiang MG, Guan SC, and Zhang LQ were analysis and interpretation of the data. Liu XL was the drafting of first version of the paper. Zhao HY and Zhu JY were revised it critically for intellectual content; and the final approval of the version to be published. No potential conflict of interest was reported by the authors and all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (919.5 KB)Disclosure statement
This word was approved by Changbai Mountain Nature Conservation Management Center and comply with the International Union for Conservation of Nature (IUCN) policies research involving species at risk of extinction (see Guidelines for appropriate uses of IUCN Red list data), the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. OP221050. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA906719, SRR22459652, and SAMN31936196, respectively.
Additional information
Funding
References
- Chen XC, Li QS, Li Y, Qian J, Han JP. 2015. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front Plant Sci. 6:1–9.
- Choi JE, Kim GB, Lim CE, Yu HJ, Mun JH. 2016. The complete chloroplast genome of Aconitum austrokoreense Koidz. (Ranunculaceae), an endangered endemic species in Korea. Mitochondrial DNA B Resour. 1(1):688–689.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Duan HR, Lu Y, Duan XR, Zhou XH, Wang CM, Tian FP, Wang XL, Yang HS, Cui GX. 2019. Characterization of the complete chloroplast genome of Delphinium grandiflorum L. Mitochondrial DNA B Resour. 5(1):35–36.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kim TH, Ha YH, Lee SR, Kim SC, Lee J, Kim DK, Kim HJ. 2022. The complete chloroplast genome sequence and phylogenetic position of Aconitum quelpaertense Nakai (Ranunculaceae). J Asia-Pac Biodivers.
- Kong HG, Liu WZ, Yao G, Gong W. 2017. A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): a traditional herbal medicinal genus. PeerJ. 5:e4018.
- Kong HH, Liu WZ, Yao G, Gong W. 2018. Characterization of the whole chloroplast genome of a rare and endangered species Aconitum reclinatum (Ranunculaceae) in the United States. Conserv Genet Resour. 10(2):165–168.
- Li LQ, Kadota Y. 2001. Aconitum. Vol. 6. Beijing, China/St. Louis (MO): Science Press/Missouri Botanical Garden Press; p. 149–222.
- Li Q, Li X, Qieyang R, Nima C, Dongzhi D, Duojie, Guo X, 2020. Characterization of the complete chloroplast genome of the Tangut monkshood Aconitum tanguticum (Ranunculales: Ranunculaceae). Mitochondrial DNA B Resour. 5(3):2306–2307.
- Lim CE, Kim GB, Baek S, Han SM, Yu HJ, Mun JH. 2017. The complete chloroplast genome of Aconitum chiisanense Nakai (Ranunculaceae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):75–76.
- Lim CE, Ryul BK, Lee JD, Jung KD, Noh TK, Lee BY. 2020. The complete chloroplast genome of Aconitum puchonroenicum Uyeki & Sakata (Ranunculaceae), a rare endemic species in Korea. Mitochondrial DNA Part B. 5(2):1284–1285.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu Y, Yu SH, You FM. 2020. Characterization of the complete chloroplast genome of Aconitum flavum (Ranunculaceae). Mitochondrial DNA B Resour. 5(3):2982–2983.
- Ni XS, Li J, Li YF, Zhang HZ, Duan BZ, Chen XB, Xia CL. 2022. The complete chloroplast genome of Aconitum piepunense (Ranunculaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 7(1):115–117.
- Park I, Kim WJ, Yang S, Yeo SM, Li H, Moon BC. 2017. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLOS One. 12(9):e0184257.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wang ZH, Li YQ. 2020. Characterization of the complete chloroplast genome of Aconitum pendulum (Ranunculaceae), an endemic medicinal herb. Mitochondrial DNA B Resour. 5(1):382–383.
- Xia CL, Wang MJ, Guan YH, Li J. 2022. Comparative analysis of the chloroplast genome for Aconitum species: genome structure and phylogenetic relationships. Front Genet. 13:878182.
- Xiao PG, Wang FP, Gao F, Yan LP, Chen DL, Liu Y. 2006. A pharmacophylogenetic study of Aconitum L. (Ranunculaceae) from China. Acta Phytotaxon Sin. 44(1):1–46.
- Yang MH, Xia CL, Guan YH, Zhang HZ, Chen XB, Wang Y. 2022. Sequencing and characterization of the chloroplast genome of Aconitum forrestii Stapf provide insights into phylogenetics in Aconitum. Mitochondrial DNA B Resour. 7(6):1165–1167.
- Zhang M, Luo JW, Su LJ, Ding QJ, Yin XM, Hou FX, Gao JH, Peng C. 2021. The complete chloroplast genome of Aconitum scaposum. Mitochondrial DNA B Resour. 6(8):2149–2150.
- Zhou Y. 2006. The research on the sequences of preferential protection of the rare and endangered plants in Changbai mountain. Forest Res. 19:740–749.
- Zhou Y. 2009. Chang Bai mountains plant resources China. Beijing: China Forestry Press; p. 290.