Abstract
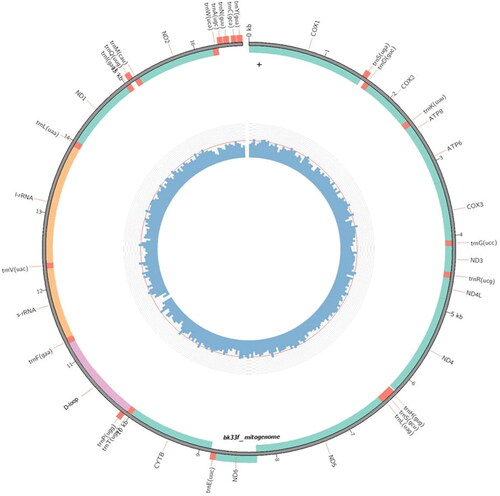
In this study, we report the nearly complete mitochondrial sequence of Batagur affinis affinis. The assembled mitogenome consists of 13 PCGs, 22 tRNA genes, two rRNAs and one near-complete D-loop region. Of the annotated genes, the ND6 subunit gene and eight tRNA genes were encoded on the L-strand, while the remaining genes were dispersed on the H-strand. Except for CO1, which has a GTG start codon, all protein-coding genes begin with ATG. The mitogenome has been deposited in NCBI GenBank under the accession number OQ409915. Phylogenetic tree analysis based on publicly available mitogenomes indicate the sister grouping of B. affinis affinis with B. kachuga.
Introduction
The Southern River Terrapin, locally known as Tuntung Sungai, is a member of the genus Batagur, a family of Geoemydidae and the class Reptilia. The six species in the genus Batagur are the Three-striped Roofed Turtle (Batagur dhongoka), Northern River Terrapin (Batagur baska), Red-crowned Roofed Turtle (Batagur kachuga), Burmese Roofed Turtle (Batagur trivittata), Southern River Terrapin (Batagur affinis ssp.), and Painted Terrapin (Batagur borneoensis) (Uwe and Havas Citation2007). Five of the six Batagur species, including the study’s focal species, are on the list of the top 25 most endangered turtles, except for Batagur dhongoka (Turtle Conservation Coalition Citation2011), which is classified as critically endangered on the IUCN 2000 Red List (Horne et al. Citation2016) and is close to extinction. Batagur affinis ssp. (Cantor Citation1847) is among 24 species of turtles found in Malaysia (Mohd-Salleh et al. Citation2022), whose closest Southeast Asian cousins are B. baska and B. kachuga.
According to Praschag et al. (Citation2007), B. baska was made up of at least two heritably different species: populations of B. affinis affinis in the Kedah and Perak river systems on the west coast of Peninsular Malaysia. However, Salleh et al. (Citation2022) and Salleh et al. (Citation2023) recently stated B. affinis edwardmolli is also found in Kedah, Malaysia. On the other hand, the populations in the Terengganu river basin were found to be B. affinis edwardmolli.
Materials and methods
In this study, we used the genome skimming approach to recover the near-complete mitochondrial genome of B. affinis affinis (). The blood sample was collected from a female individual at the Southern River Terrapin Conservation Center, Bota Kanan, Perak, Malaysia (4.3489° N, 100.8802° E) and stored at −20 °C in the Laboratory of Breeding Genetics at Universiti Putra Malaysia. The specimen is kept in the Depository Museum, Department of Aquaculture, Faculty of Agriculture, Universiti Putra Malaysia (Dr. Zafri Hassan, +60397694932, [email protected]). The specimen has a voucher number of JAQ/BA/00001.
Figure 1. Represent an adult Southern River Terrapin (B. affinis affinis) from the Southern River Terrapin Breeding Center in Bota Kanan, Perak, Malaysia (own photo).

Total genomic DNA was extracted from approximately 2 mL of the blood sample following noninvasive procedures developed by Salleh and Esa (Citation2022) and used for DNA extraction using the Qiagen Qiamp Minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA was sheared to 350 bp using a Bioruptor, followed by library preparation using the NEBUltra II Library Preparation Kit according to the manufacturer’s instructions. The constructed library was quantified with TapeStation (Agilent, Santa Clara, CA), followed by sequencing on a NovaSEQ6000 (Illumina, San Diego, CA) using a run configuration of 2 × 150 bp to generate approximately 5 GB of data. Raw paired-end reads were filtered with fastp (default setting) followed by de novo assembly using MEGAHIT v1.2.9. Identification of mitochondrial-derived contig(s) from the de novo assembly and mitogenome annotation used MitoZ v3.4.
Results
The first mitochondrial genome sequence from B. affinis affinis (specimen ID: BK33F) was submitted to GenBank and assigned the accession number OQ409915 (). The assembled mitogenome length was 16,526 bp consisting of 13 conventional vertebrate protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a near-complete control region (D-loop) with no notable variations from the standard vertebrate mtDNA gene organization (Li et al. Citation2017; Tang et al. Citation2019).
Discussion and conclusion
The mitogenome is composed of A (32.4%), T (25.3%), C (29.5%), and G (12.4%), in that order. A + T indicated a typical mitogenome sequence (61.0%). CO1 is the only protein-coding gene that begins with a start codon other than ATG. Most protein-coding genes are terminated by the stop codon TAA. Nine protein-coding genes end with complete stop codons (AGG, TAA, and TAG). Still, the remaining three genes end with T or TA as partial stop codons, which are presumed completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). Except for the ND6 gene and eight tRNA genes, most B. affinis affinis mitochondrial genes are encoded on the H-strand. The trnCgca gene was the shortest among the mitochondrial protein-coding genes, while the ND5 gene was the longest. The 12S and 16S ribosomal RNAs have lengths of 965 and 1595 base pairs, respectively.
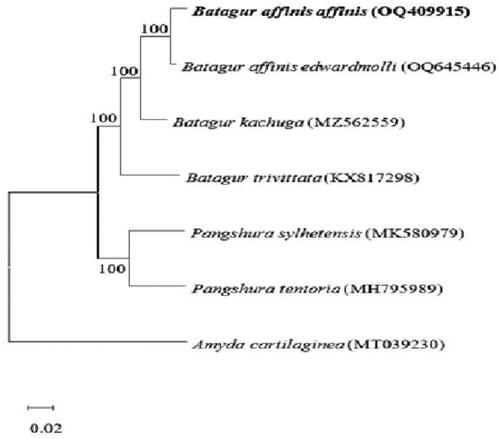
Surprisingly, only two Batagur mitogenomes (B. kachuga and B. trivittata) have been reported in GenBank (https://www.ncbi.nlm.nih.gov/genbank) to date, despite the vast number of species in this genus. depicts the construction of a neighbor-joining (NJ) tree (Saitou and Nei Citation1987) using the dataset and MEGA X (Kumar et al. Citation2018). According to the cluster in the phylogenetic tree, B. affinis affinis and B. kachuga have the closest evolutionary ties. In contrast to the conventional CO1 or CytB partial gene sequence, the full mitogenome offers a much greater number of variable sites dramatic increase in the number of variable sites. It thus will be helpful in delineating the evolutionary relationship of Batagur spp. in the future.
Figure 3. Neighbor-joining tree showing the genetic related of Batagur spp. The evolutionary history was inferred using the Neighbour-Joining method (NJ) (Saitou and Nei Citation1987). the bootstrap consensus tree inferred from 1000 replicates (Felsenstein Citation1985) is taken to represent the evolutionary history of the taxa analyzed (Felsenstein Citation1985). Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein Citation1985). the evolutionary distances were computed using the Kimura 2-parameter method (Kimura Citation1980) and are in the units of the number of base substitutions per site. This analysis involved four nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). there was a total of 11391 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al. Citation2018). the following sequences were used: Batagur affinis edwardmolli OQ645446 (Salleh et al. Citation2023), Batagur kachuga MZ562559 (Das et al. Citation2021), Batagur trivittata KX817298 (Feng et al. Citation2017), Pangshura sylhetensis MK580979 (Kundu et al. Citation2020), Pangshura tentoria MH795989 (Kundu et al. Citation2019), and Amyda cartilaginea MT039230 (Cui et al. Citation2020).
Author contributions
Mohd Hairul Mohd Salleh: conceptualization, methodology, formal analysis, investigation, and writing—original draft. Yuzine Esa: conceptualization, writing (review and editing), funding acquisition, resources, and advisor. Han Ming Gan: formal analysis, investigation, and report (review and editing). All authors agreed to be accountable for all aspects of the work and approved the final draught to be published.
Ethics statement
The Department of Wildlife and National Parks, Peninsular Malaysia, permitted and approved the study (B-00335-16-20) to comply with the International Union for Conservation of Nature (IUCN) policies for research involving species at risk of extinction.
Acknowledgments
We want to express our gratitude to the Laboratory of Breeding Genetics, Department of Aquaculture, Faculty of Agriculture, Universiti Putra Malaysia, for the facilities and support of this research project. We also want to thank the Turtle Conservation Society of Malaysia and the Department of Wildlife and National Parks, Peninsular Malaysia, for working together.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/under the accession number OQ409915. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA767629, SRR22444522, and SAMN31872828, respectively.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465. doi: 10.1038/290457a0.
- Cantor T. 1847. Catalog of reptiles inhabiting the Malayan peninsula and islands. Journal of the Asiatic Society of Bengal. 16:607–656, 897–952, 1026–1078.
- Cui L, Rao D, Zhang M. 2020. The complete mitochondrial genome of Amyda cartilaginea (Testudines: Trionychidae). Mitochondrial DNA Part B. 5(3):3652–3654. doi: 10.1080/23802359.2020.1832595.
- Das SP, Krishnan R, Majhi A, Sunil M, Tejwan Y, Srivastava A, Gadnayak A. 2021. Batagur kachuga isolate BK1 mitochondrion, complete genome. [accessed from] https://www.ncbi.nlm.nih.gov/nucleotide/MZ562559.1?report=genbank&log$=nucltop&blast_rank=2&RID=RT0UBXHV01N.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x.
- Feng L, Yang J, Zhang YP, Zhao GF. 2017. The complete mitochondrial genome of the Burmese roofed turtle (Batagur trivittata) (Testudines: Geoemydidae). Conservation Genet Resour. 9(1):95–97. doi: 10.1007/s12686-016-0629-5.
- Horne BD, Chan EH, Platt SG, Moll EO. 2016. Batagur affinis. The IUCN Red List of Threatened Species [accessed 2017 Aug 13]. doi: 10.2305/IUCN.UK.2016-.1.RLTS.T170501A1315041.en.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. doi: 10.1007/BF01731581.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Kundu S, Kumar V, Tyagi K, Chandra K. 2020. The complete mitochondrial genome of the endangered Assam Roofed Turtle, Pangshura sylhetensis (Testudines: Geoemydidae): Genomic features and phylogeny. PLoS One. 15(4):e0225233. doi: 10.1371/journal.pone.0225233.
- Kundu S, Kumar V, Tyagi K, Chakraborty R, Chandra K. 2019. The first complete mitochondrial genome of the Indian Tent Turtle, Pangshura tentoria (Testudines: Geoemydidae): characterization and comparative analysis. Ecol Evol. 9(18):10854–10868. doi: 10.1002/ece3.5606.
- Li J, Lu Y, Zan J, Nie L. 2017. Complete mitochondrial genome of the Cyclemys pulchristriata (Chelonia: Geoemydidae). Mitochondrial DNA B Resour. 2(2):403–404. doi: 10.1080/23802359.2017.1347836.
- Mohd-Salleh MH, Esa Y, Salleh SM, Mohd Sah SA. 2022. Turtles in Malaysia: a Review of Conservation Status and a Call for Research. Animals. 12(17):2184. doi: 10.3390/ani12172184.
- Praschag P, Hundsdörfer AK, Fritz U. 2007. Phylogeny and taxonomy of endangered South and Southeast Asian freshwater turtles elucidated by mtDNA sequence variation (Testudines: geoemydidae: batagur, Callagur, Hardella, Kachuga, Pangshura). Zool Scripta. 36(5):429–442. doi: 10.1111/j.1463-6409.2007.00293.x.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Salleh MHM, Esa Y, Mohamed R, Pelf-Nyok C, Ismail HM. 2022. Saving Freshwater Turtle – Batagur affinis. In: Shanbhag S., editors. Emerging trends in science, social science, engineering and management-a multidisciplinary approach. India: Research Circle & International Institute of Knowledge and Research; p. 296–307.
- Salleh MHM, Esa Y, Pau S-SN. 2023. Conservation genetics of the critically endangered Southern River Terrapin (Batagur affinis) in Malaysia: genetic diversity and novel subspecies distribution ranges. Biology. 12(4):520. doi: 10.3390/biology12040520.
- Salleh MHM, Esa Y. 2022. Minimally invasive blood collection techniques as a source of gDNA for genetic studies on turtles and tortoises. Int J Aquatic Biol. 10(2):145–150.
- Tang XS, Yang DC, Peng LF, Weng SY, Huang S. 2019. The complete mitochondrial genome of the critically endangered Cuora mccordi (Reptilia: geoemydidae). Mitochondrial DNA Part B. 4(1):480–481. doi: 10.1080/23802359.2018.1536493.
- Turtle Conservation Coalition [Rhodin AGJ, Walde AD, Horne BD, Van Dijk PP, Blanck T, Hudson R.(eds)]. 2011. Turtles in trouble: the World’s 25+ most endangered Tortoises and Freshwater Turtles—2011. Lunenburg, MA: IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Turtle Conservation Fund, Turtle Survival Alliance, Turtle Conservancy, Chelonian Research Foundation, Conservation International, Wildlife Conservation Society, and San Diego Zoo Global, p. 54. pp. (2) (PDF) Turtles in Trouble: the World’s 25+ Most Endangered Tortoises and Freshwater Turtles—2011.
- Uwe F, Havas P. 2007. Checklist of chelonians of the World. VZ. 57(2):149–368. doi: 10.3897/vz.57.e30895.