Abstract
Primula amethystina subsp. argutidens (Franchet) W. W. Smith & H. R. Fletcher (1942) is a blooming plant of the family Primulaceae. Here, we sequenced, assembled, and annotated the complete chloroplast (cp) genome of P. amethystina subsp. argutidens. The cp genome of P. amethystina subsp. argutidens is 151,560 bp in length with a GC content of 37%. The assembled genome has a typical quadripartite structure, containing a large single-copy (LSC) region of 83,516 bp, a small single-copy (SSC) region of 17,692 bp, and a pair of inverted repeat (IR) regions of 25,176 bp. The cp genome contains 115 unique genes, including 81 protein-coding genes, four rRNA genes, and 30 tRNA genes. Phylogenetic analysis showed that P. amethystina subsp. argutidens was closely related to P. amethystina.
Introduction
Primula amethystina subsp. argutidens (Franchet) W. W. Smith & H. R. Fletcher (Smith and Fletcher Citation1942) is a blooming plant of the family Primulaceae (). In Flora of China, P. amethystina subsp. argutidens is classified as a subspecies of P. amethystina (Hu and Kelso Citation1996). This subspecies is a perennial herb with rich violet-blue flowers and deeply serrated leaves, mainly distributed in alpine grasslands at an altitude of 3500–5000 m in the western Sichuan Province of China (Hu and Kelso Citation1996). Compared with P. amethystina and P. amethystina subsp. brevifolia, P. amethystina subsp. argutidens has a more petite stem body, deeper-toothed leaf margins, shorter pedicels, and a profoundly recessed corolla lobe end (Hu and Kelso Citation1996). Although cp genome is widely used in phylogenetic research and molecular identification in many families of angiosperm, plastome phylogenomic studies of the phylogenetic relationship between P. amethystina subsp. argutidens and P. amethystina are lacking (Huang et al. Citation2017; Bi et al. Citation2018). Here, we report the first complete cp genome of Primula amethystina subsp. argutidens to provide a plastome database for further phylogenetic analysis.
Materials and methods
In this study, P. amethystina subsp. argutidens was sampled from Shangri-la of Yunnan Province, China (27°37′11″N, 99°38′29″E). Voucher specimens (HY-22) were deposited in the Herbarium of Yunnan Normal University (YNUB, Website: https://life.ynnu.edu.cn/, Contact: Jian-Lin Hang, Email: [email protected]). Total genomic DNA was extracted using a modified CTAB method (Porebski et al. Citation1997). The fragmented genomic DNA was used to construct short-insert libraries for Illumina paired-end (PE) sequencing on the Illumina Hiseq X Ten sequencer. We obtained 17,593,572 filtered reads and then assembled the plastid genome using the software NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017) with P. amethystina as the reference genome (GenBank accession no. NC_053577). Then, we used Geneious V2020.1.1 software (Kearse et al. Citation2012) to annotate the cp genome of P. amethystina subsp. argutidens using P. amethystina (Wang et al. Citation2021) as a reference and then used Primula bulleyana (Chen, Zhang et al. Citation2019) as a reference for modification and correction. The annotated cp genome sequences of P. amethystina subsp. argutidens were deposited in the GenBank database under the accession no. ON416902. The circular Primula amethystina subsp. argutidens genomic map and schematic map of the cis and trans splicing genes were drawn using CPGView (Liu et al. Citation2023, http://www.1kmpg.cn/cpgview/). To verify the accuracy of the assembly, we mapped clean reads to the assembled chloroplast (cp) genomes to assess the depth of coverage (Figure S1). We used DnaSP6.12 to perform a sliding window analysis of nucleotide diversity to evaluate the divergence of the four Sect. Amethystina cp genome sequences (Librado and Rozas Citation2009).
In order to further determine the phylogenetic relationship of P. amethystina subsp. argutidens within the genus Primula, the multiple sequence alignment of the complete cp DNA of 47 species from the Primulaceae was analyzed by MAFFT (Katoh and Standley Citation2013). Four Lysimachia species, four Androsace species, and Glaux maritima were chosen as outgroups for constructing a phylogenetic tree. The maximum-likelihood (ML) tree was constructed using IQ-TREE v1.6.10 (Nguyen et al. Citation2015), and the best-fit model according to the Bayesian information criterion (BIC) was TVM + F + R3 (Kalyaanamoorthy et al. Citation2017). Branch supports were tested using ultrafast bootstrap (UFBoot) (Hoang et al. Citation2018) and SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al. Citation2010) with 10,000 replicates.
Results
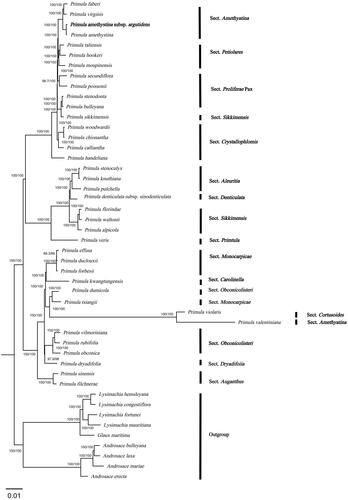
The complete cp genome of P. amethystina subsp. argutidens was 151,560 bp in length with an overall GC content of 37% and an average coverage of 1789.6 (, Figure S1). The assembled genome has a typical quadripartite structure, containing a large single-copy (LSC) region of 83,516 bp and a small single-copy (SSC) region of 17,692 bp, which are separated by a pair of inverted repeat (IR) regions of 25,176 bp. The cp genome contains 134 genes, including 89 protein-coding genes, eight rRNA genes, and 37 tRNA genes, of which 81 CDS genes, four tRNA genes, and 30 rRNA genes are unique, respectively. Totally, 11 cis-splicing genes including rps16, atpF, rpoC1, pafl, clpP, petB, petD, rpl16, rpl2, ndhB, and ndhA (Figure S2A), and one trans-splicing genes rps12 (Figure S2B) were detected. The cp genomes of four species of Sect. Amethystina were compared to screen single-nucleotide polymorphisms (SNPs) with higher nucleotide diversity value. As a result, the four most variable regions were rpl2-trnH-GUG, trnH-GUG-psbA, trnK-UUU-rps16, and rpoB-trnC-GCA of which three are located in the LSC region and one is in the SSC region. Partial sequences of trnH-GUG, psbA, rps16, rpoB, and ycf1 also have rich variable sites (Table S1, Figure S3). The phylogenetic tree demonstrated that P. amethystina subsp. argutidens and P. amethystina formed a robust monophyletic clade, which was sister to each other ().
Figure 2. Genomic map of overall features of Primula amethystina subsp. argutidens chloroplast genome, generated by CPGview (http://www.1kmpg.cn/cpgview/). The species’ name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
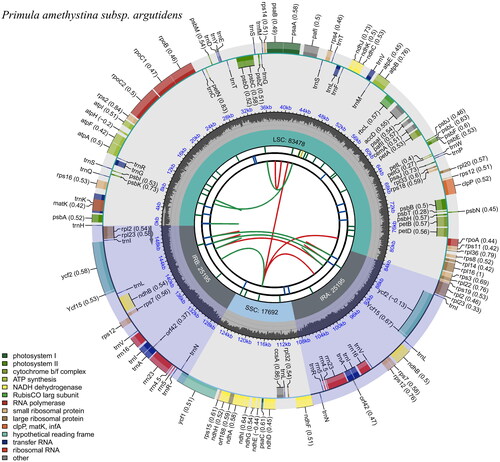
Figure 3. In the ML phylogenetic tree of P. amethystina subsp. argutidens and 46 Primulaceae species based on chloroplast genomes. The best-fit model according to the Bayesian information criterion (BIC) was TVM + F + R3. Branch supports were tested using ultrafast bootstrap (UFBoot) and SH-like approximate likelihood ratio test (SH-aLRT) with 10,000 replicates. P. amethystina subsp. argutidens is marked in bold. The following sequences were used: Primula faberi NC_053576 (Wang et al. Citation2021), Primula virginis NC_053581 (Wang et al. Citation2021), Primula amethystina NC_053577 (Wang et al. Citation2021), Primula taliensis NC_053601 (Wang et al. Citation2021), Primula hookeri NC_053593 (Wang et al. Citation2021), Primula moupinensis NC_050244, Primula secundiflora NC 053585 (Wang et al. Citation2021), Primula poissonii NC_024543 (Yang et al. Citation2014), Primula stenodonta NC 034677 (Zhang et al. Citation2017), Primula bulleyana NC_046947 (Chen, Zhang et al. Citation2019), Primula sikkimensis NC_050243, Primula woodwardii NC 039349 (Ren et al. Citation2018), Primula chionantha NC_053583 (Wang et al. Citation2021), Primula calliantha MZ054238 (Yang et al. Citation2021), Primula handeliana NC_039348 (Ren et al. Citation2018), Primula stenocalyx NC_058249 (Guo, Ma, Li et al. Citation2021), Primula knuthiana NC_039350 (Ren et al. Citation2018), Primula pulchella NC_050246 (Zhang et al. Citation2017), Primula denticulata subsp. sinodenticulata NC_050247, Primula florindae NC_053579 (Wang et al. Citation2021), Primula waltonii NC_058808, Primula alpicola NC_053588 (Wang et al. Citation2021), Primula veris NC_031428 (Zhou et al. Citation2016), Primula effusa NC_058259 (Zhong et al. Citation2019), Primula duclouxii NC_058263 (Zhong et al. Citation2019), Primula forbesii NC_061696, Primula kwangtungensis NC_034371 (Zhang et al. Citation2016), Primula dumicola NC_058257 (Zhong et al. Citation2019), Primula tsiangii NC_046755 (Chen, Yan et al. Citation2019), Primula violaris NC_050848 (Chai et al. Citation2021), Primula valentiniana NC_061669, Primula vilmoriniana NC_058258 (Zhong et al. Citation2019), Primula rubifolia NC_058261 (Zhong et al. Citation2019), Primula obconica NC_046415 (Zhang et al. Citation2019), Primula dryadifolia NC_053596 (Wang et al. Citation2021), Primula sinensis NC_030609 (Liu et al. Citation2016), Primula filchnerae NC_051972 (Lu et al. Citation2020), Lysimachia hemsleyana NC_052863 (Ying et al. Citation2019), Lysimachia congestiflora NC_045275 (Li et al. Citation2019), Lysimachia fortunei NC_052863, Lysimachia mauritiana NC_060700 (Lee et al. Citation2022), Glaux maritima NC_059901 (Liu et al. Citation2021), Androsace bulleyana NC_034641, Androsace laxa (Ren et al. Citation2018), Androsace mariae NC_051991 (Guo, Ma, Xie et al. Citation2021), and Androsace erecta NC 057637.
Conclusions
Our result supported well the subspecies of P. amethystina subsp. argutidens. The complete cp genome sequence of P. amethystina subsp. argutidens provided a valuable genomic database for further studies on the evolutionary history and phylogenetic relationship of Primula species.
Ethical approval
This article does not contain any studies with human participants or animals performed by any authors. The species described in this paper are not endangered, protected, or personally owned. The plant material was collected in accordance with guidelines provided by the authors’ institution (School of Life Sciences, Yunnan Normal University) and national regulations.
Author contributions
Yuan Huang and Shu-Bao Wang conceived and designed this study. Shu-Bao Wang, Yun-Qi Liu, and Li Zhang conducted an analysis. Shu-Bao Wang and Rui Li contributed to the analytical methods. Shu-Bao Wang wrote the manuscript. Yuan Huang revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Word (454.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the study’s findings are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/ under accession no. ON416902. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA834988, SRR19175835, and SAMN28088488, respectively.
Additional information
Funding
References
- Bi Y, Zhang MF, Xue J, Dong R, Du Y-P, Zhang X-H. 2018. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. 8(1):1184. doi:10.1038/s41598-018-19591-9.
- Chai W, Li H, Zhang C, Zhao B, Xia P. 2021. The complete chloroplast genome of the ornamental plant Primula violaris (Primulaceae). Mitochondrial DNA B. 6(12):3429–3430. doi:10.1080/23802359.2021.2002206.
- Chen S, Yan X, Hao G, Xu Y. 2019. The complete chloroplast genome of Primula tsiangii W. W. Smith (Primulaceae): a karst endemic primrose in Southwest China. Mitochondrial DNA B. 4(2):2627–2628. doi:10.1080/23802359.2019.1642160.
- Chen X, Zhang L, Li WQ, Huang Y, Wu Z. 2019. The complete chloroplast genome of Primula bulleyana, a popular ornamental species. Mitochondrial DNA B. 4(2):3673–3674. doi:10.1080/23802359.2019.1678436.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi:10.1093/sysbio/syq010.
- Guo Y, Ma L, Li J. 2021. The complete chloroplast genome and phylogenetic analysis of Primula stenocalyx Maxim. Mitochondrial DNA B. 6(11):3231–3232. doi:10.1080/23802359.2021.1991245.
- Guo Y, Ma R, Xie J, Liu Q, Zhu Y, Yang W, Gao L, Ma Y. 2021. The complete chloroplast genome of Androsace mariae. Mitochondrial DNA B. 6(2):376–377. doi:10.1080/23802359.2020.1866463.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi:10.1093/molbev/msx281.
- Hu Q, Kelso S. 1996. Primulaceae. In: Wu CY, Raven PH, editors. Flora of China. Vol. 15. Beijing and St. Louis: Science Press and Missouri Botanical Garden; p. 139.
- Huang Y, Wang J, Yang Y, Fan C, Chen J. 2017. Phylogenomic analysis and dynamic evolution of chloroplast genomes in Salicaceae. Front Plant Sci. 8:1050. doi:10.3389/fpls.2017.01050.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Lee Y, Yun N, Kang J, Choi S, Paik J-H. 2022. The complete chloroplast genome of the medicinal plant Lysimachia mauritiana (Lamarck, 1792). Mitochondrial DNA B. 7(3):554–555. doi:10.1080/23802359.2022.2054738.
- Li HL, Cheng XL, Chen Y, Tan F-L. 2019. Complete plastome sequence of Lysimachia congestiflora Hemsl. a medicinal and ornamental species in Southern China. Mitochondrial DNA B. 4(2):2316–2317. doi:10.1080/23802359.2019.1627952.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11):1451–1452. doi:10.1093/bioinformatics/btp187.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23:1–11.
- Liu TJ, Zhang CY, Yan HF, Zhang L, Ge X-J, Hao G. 2016. Complete plastid genome sequence of Primula sinensis (Primulaceae): structure comparison, sequence variation and evidence for accD transfer to nucleus. PeerJ. 4:e2101. doi:10.7717/peerj.2101.
- Liu YQ, Chen X, Zhang L, Huang Y. 2021. The complete chloroplast genome of Glaux maritima, a monotypic species. Mitochondrial DNA B. 6(8):2137–2138. doi:10.1080/23802359.2021.1944370.
- Lu Y, Liu P-L, Sun Y-F, Li S-F. 2020. The complete chloroplast genome sequence of Primula filchnerae Knuth (Primulaceae), an endangered species in China. Mitochondrial DNA B Resour. 5(3):2047–2048. doi:10.1080/23802359.2020.1750991.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. doi:10.1007/BF02772108.
- Ren T, Yang Y, Zhou T, Liu ZL. 2018. Comparative plastid genomes of Primula species: sequence divergence and phylogenetic relationships. Int J Mol Sci. 19(4):1050. doi:10.3390/ijms19041050.
- Smith WW, Fletcher HR. 1942. The genus Primula: section Amethystina. Trans Bot Soc Edinburgh. 33(3):209–226. doi:10.1080/13594864209441382.
- Wang X-J, Barrett SCH, Zhong L, Wu Z-K, Li D-Z, Wang H, Zhou W. 2021. The genomic selfing syndrome accompanies the evolutionary breakdown of heterostyly. Mol Biol Evol. 38(1):168–180. doi:10.1093/molbev/msaa199.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Yang X, Huang Y, Li Z, Chen J. 2021. Complete plastid genome of Primula calliantha Franch. (Primulaceae): an alpine ornamental plant endemic to Hengduan Mountain, China. Mitochondrial DNA B Resour. 6(9):2643–2645. doi:10.1080/23802359.2021.1963868.
- Ying Z, Wang Q, Yu S, Liao G, Ge Y, Cheng R. 2019. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Lysimachia hemsleyana. Mitochondrial DNA B Resour. 4(2):3878–3879. doi:10.1080/23802359.2019.1688115.
- Zhang C, Yuan X, Yang T, Yan H, Liu T. 2019. The complete chloroplast genome of Primula obconica (Primulaceae). Mitochondrial DNA B Resour. 4(2):2189–2190. doi:10.1080/23802359.2019.1623101.
- Zhang CY, Liu TJ, Xu Y, Yan H-f. 2016. Characterization of the whole chloroplast genome of an endangered species Primula kwangtungensis (Primulaceae). Conserv Genet Resour. 9(1):1–3.
- Zhang C-Y, Liu T-J, Yan H-F, Ge X-J, Hao G. 2017. The complete chloroplast genome of a rare candelabra primrose Primula stenodonta (Primulaceae). Conserv Genet Resour. 9(1):123–125. doi:10.1007/s12686-016-0637-5.
- Zhong L, Barrett SCH, Wang X-J, Wu Z-K, Sun H-Y, Li D-Z, Wang H, Zhou W. 2019. Phylogenomic analysis reveals multiple evolutionary origins of selfing from outcrossing in a lineage of heterostylous plants. New Phytol. 224(3):1290–1303. doi:10.1111/nph.15905.
- Zhou T, Zhao J, Chen C, Meng X, Zhao G. 2016. Characterization of the complete chloroplast genome sequence of Primula veris (Ericales: Primulaceae). Conserv Genet Resour. 8(4):455–458. doi:10.1007/s12686-016-0595-y.

