Abstract
Rana coreana is a brown frog species native to the Korean Peninsula. We characterized the complete mitochondrial genome of the species. The mitochondrial genome sequence of R. coreana is 22,262 bp and comprises 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two control regions (CRs). The CR duplication and gene organization were identical to those observed in Rana kunyuensis and Rana amurensis. A total of 13 protein-coding genes were used to examine the phylogenetic relationships between this species and the genus Rana. R. coreana living on the Korean Peninsula, formed a cluster with R. kunyuensis and R. amurensis, with R. coreana showing the closest phylogenetic affinity for R. kunyuensis.
Introduction
Rana coreana (Okada, 1928) is one of three brown frog species found in Korea, along with Rana uenoi and Rana huanrenensis (Kim et al. Citation2002; Song et al. Citation2006; Yang et al. Citation2017). R. coreana is also known as Rana amurensis; however, according to Song et al. (Citation2006), it demonstrates morphological and genetic differences from R. amurensis. R. coreana differs from other brown frogs in having a continuous white line along the upper lip and distinct dark speckling extending from behind the eardrums to the snout tip (). R. coreana is an endemic species restricted to Korea. In 2015, R. kunyuensis, which inhabits the Kunyu Mountains of the Shandong Peninsula in China, was pronounced as a junior synonym of R. coreana (Zhou et al. Citation2015). The mitochondrial genome sequence of R. kunyuensis has been reported by Li, Yin, et al. (Citation2016). However, there is no complete mitochondrial genome of R. coreana. This study aimed to present the complete mitochondrial genome, which contributes to the genetic diversity of brown frogs.
Figure 1. Rana coreana specimen. Rana coreana is a brown frog found in South Korea. This species differs from other brown frogs in having a continuous white line along its upper lip, as shown in the figure. Photograph was taken of National Institute of Biological Resources (NIBR) immersion specimens (NIBR AM0000000711).

Materials
The R. coreana specimen was collected from Boeun-gun, Chungcheongbuk-do, Republic of Korea (N32°34′15.46″, E124°52′24.38″). Tissue biopsy was performed at the National Institute of Biological Resources (NIBR, Incheon, Republic of Korea; https://www.nibr.go.kr/cmn/main/enMain.do; contact Jung A Kim ([email protected]) under voucher number NIBRGR0000654507). The sampled frog was released immediately after sampling and bleeding was stopped using an alcohol swab.
Methods
DNA was extracted from 2 mm frog toe samples using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) per the manufacturer’s protocol. The MGIEasy DNA Library Prep Kit was obtained from MGI (Shenzhen, China), and next-generation sequencing (NGS) reads were generated from the DNBSEQ-T7RS (MGI, Shenzhen, China) platform with a read length of 150 bp. The mitochondrial genome was assembled using MitoZ (v.2.3) (Meng et al. Citation2019) and two gaps were assembled using long-PCR analysis and Sanger sequencing (Table S1 and Figure S1). Sanger sequencing was used to build an assembly using CAP3 (version date 02/10/15) (Figure S2) (Huang and Madan Citation1999). Small gaps in the assembly were re-mapped using MITOBim (v.1.9.1) (Hahn et al. Citation2013) and error correction was conducted using PILON (v.1.24) (Walker et al. Citation2014). The final assembly was manually curated (Table S2). Gene prediction of R. coreana mitogenome was performed using GeSeq (Tillich et al. Citation2017) and MITOS (Bernt et al. Citation2013). A mitogenome map of R. coreana was generated using CGView (https://proksee.ca) (Grant and Stothard Citation2008). We constructed a phylogenetic tree for the 16 Rana species using 13 protein-coding genes with the R. sylvatica as an outgroup. To construct the phylogenetic tree, we used the neighbor-joining algorithm with the Poisson model in MEGA11 (Tamura et al. Citation2021) and conducted bootstrap analysis with 500 replicates to assess the tree robustness. Tree information was visualized using FigTree (ver. 1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/).
Results
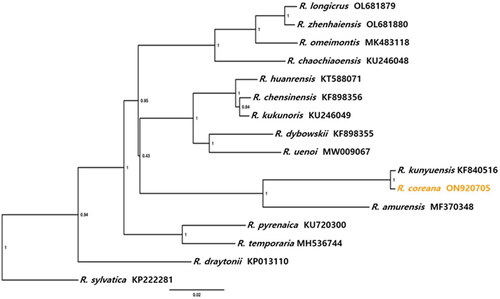
The entire mitochondrial genome (mitogenome) sequence of R. coreana (GenBank accession: ON920705) was 22,262 bp in length, including 13 protein-coding genes, two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes (, Table S2). The mitogenome contained two control regions (CRs) that increased the genome size of R. coreana (Figure S3). Phylogenetic relationships among the mitogenomes of 15 East Asian Rana species revealed the closest affinity between R. coreana and R. kunyuensis ().
Figure 2. Mitochondrial genome map of Rana coreana. Mitochondrial genome map. Genes located outside the circle are transcribed in the heavy-strand direction, whereas genes inside the circle are transcribed in the light-strand direction. The genomic coordinates of R. coreana mitochondrial genes are summarized in Table S2.
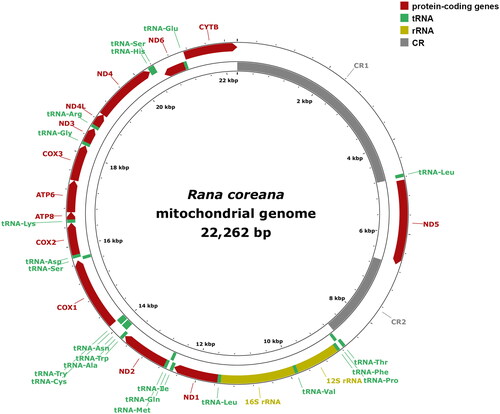
Figure 3. Phylogenetic tree relationship. The phylogenetic tree of 16 species in Rana based on mitochondrial genome, including R. longicrus (Chen et al. Citation2022), R. zhenhalensis (Chen et al. Citation2022), R. omeimontis (Jiang et al. Citation2020), R. chaochiaoensis, R. huanrensis (Dong et al. Citation2016), R. chensinensis (Li, Lei, et al. Citation2016), R. kukunoris (Wang et al. Citation2020), R. dybowskii (Li, Lei, et al. Citation2016), R. uenoi (Suk et al. Citation2021), R. kunyuensis (Li, Yin, et al. Citation2016), R. amurensis (Liu et al. Citation2017), R. pyrenaica (Peso-Fernãndez et al. Citation2016), R. temporaria (Chen Citation2018), R. draytonii (Li, Lei, et al. Citation2016), and R. sylvatica (Ni et al. Citation2016). R. coreana (orange) is available under NCBI GenBank accession number ON920705. The GenBank accession numbers for the sequences are indicated next to the species names. Bootstrap values right to the nodes.
Discussion and conclusions
We assembled the mitogenome of R. coreana, which was 22,262 bp in length, with CR duplication ( and Figure S3). It is considerably larger than the standard vertebrate mitogenomes (Formenti et al. Citation2021). Moreover, the R. coreana mitogenome shares CR duplication and rearrangement with two other East Asian brown frogs, R. kunyuensis and R. amurensis (Figure S4) (Li, Yin, et al. Citation2016; Liu et al. Citation2017). These three taxa form a separate clade in the phylogenetic tree obtained for the 16 Rana species using 13 protein-coding genes (). This finding supports both the common ancestry and evolutionary conservation of CR duplication and the unique gene order (Chen et al. Citation2022). The sister relationship between R. coreana and R. kunyuensis and the small nucleotide divergence of their mtDNAs suggest that nuclear genes should be studied to verify their taxonomic status. These findings support those of previous studies and highlight the evolutionary conservation of this genomic change in closely related Rana species. Overall, our study contributes to the understanding of the genetic diversity and evolution of the genus Rana and provides valuable insights for future studies in this field.
Author contributions
All authors contributed to the conception and design of this study. M. M. and J. A. K. conducted material preparation and species identification. J. K., J. C., and M. S. K. conducted the bioinformatics data processing and analyses. J. B. and J. A. obtained funding, conceptualized the study, and provided supervision. J. K., J. A. K., and M. M. wrote and revised the first draft of the manuscript. All authors commented on the previous versions of the manuscript. All the authors have read and approved the final version of the manuscript.
Ethical approval
Sample collection protocols were approved by the Institutional Animal Care and Use Committee (approval number: NIBR IACUC 20220001).
Supplemental Material
Download MS Word (632.1 KB)Acknowledgements
We thank Jaesu Bhak for editing the manuscript and Daecheol Jeong for sample collection and species identification. Genetic samples for this study were collected with legal permits obtained from the Ministry of Environment, Republic of Korea.
Disclosure statement
J. B. is the CEO of Clinomics, Inc. The authors declare that they have no competing financial interests or personal relationships that could influence the work reported in this study.
Data availability statement
Genome sequencing data supporting the findings of this study are available in the NCBI GenBank database at ncbi.nlm.nih.gov. The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. ON920705. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA909043, SRR22542064, and SAMN32064903, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chen JJ. 2018. The complete mitochondrial genome of common terrestrial frog (Rana temporaria). Mitochondrial DNA B Resour. 3(2):978–979. doi:10.1080/23802359.2018.1507649.
- Chen W, Qian W, Miao K, Qian R, Yuan S, Liu W, Dai J, Hu C, Chang Q. 2022. Comparative mitogenomics of true frogs (Ranidae, Anura), and its implications for the phylogeny and evolutionary history of Rana. Animals. 12(10):1250. doi:10.3390/ani12101250.
- Dong B, Zhou Y, Yang B. 2016. The complete mitochondrial genome of the Rana huanrensis (Anura: Ranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4551–4552. doi:10.3109/19401736.2015.1101558.
- Formenti G, Rhie A, Balacco J, Haase B, Mountcastle J, Fedrigo O, Brown S, Capodiferro MR, Al-Ajli FO, Ambrosini R, et al. 2021. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 22(1):120. doi:10.1186/s13059-021-02336-9.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184. doi:10.1093/nar/gkn179.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129. doi:10.1093/nar/gkt371.
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9(9):868–877. doi:10.1101/gr.9.9.868.
- Jiang L, Zhang M, Deng L, Xu Z, Shi H, Jia X, Lai Z, Ruan Q, Chen W. 2020. Characteristics of the mitochondrial genome of Rana omeimontis and related species in Ranidae: gene rearrangements and phylogenetic relationships. Ecol Evol. 10(23):12817–12837. doi:10.1002/ece3.6824.
- Kim JB, Min MS, Yang SY, Matsui M. 2002. Genetic relationships among Korean brown frog species (Anura, Ranidae), with special reference to evolutionary divergences between two allied species Rana dybowskii and R. huanrenensis. Zoolog Sci. 19(3):369–382. doi:10.2108/zsj.19.369.
- Li J, Lei G, Fu C. 2016. Complete mitochondrial genomes of two brown frogs, Rana dybowskii and Rana cf. chensinensis (Anura: Ranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):155–156. doi:10.3109/19401736.2013.878921.
- Li J, Yin W, Xia R, Lei G, Fu C. 2016. Complete mitochondrial genome of a brown frog, Rana kunyuensis (Anura: Ranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):34–35. doi:10.3109/19401736.2013.869681.
- Liu P, Wang H, Zhao W. 2017. Sequencing and analysis of the complete mitochondrial genome of Rana amurensis (Anura: Ranidae). Mitochondrial DNA B Resour. 2(2):424–425. doi:10.1080/23802359.2017.1357444.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi:10.1093/nar/gkz173.
- Ni N, Yu D, Storey KB, Zheng R, Zhang J. 2016. The complete mitochondrial genome of Lithobates sylvaticus (Anura: Ranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2460–2461. doi:10.3109/19401736.2015.1033697.
- Peso-Fernãndez M, Ponti De La Iglesia R, Ponz Segrelles G, Gonzãlez Martïnez R, Arcones Segovia A, Vieites DR. 2016. The complete mitochondrial genome of the endangered European brown frog Rana pyrenaica through RNAseq. Mitochondrial DNA B Resour. 1(1):394–396. doi:10.1080/23802359.2016.1174087.
- Song JY, Matsui M, Chung KH, Oh HS, Zhao W. 2006. Distinct specific status of the Korean brown frog, Rana amurensis coreana (Amphibia: Ranidae). Zoolog Sci. 23(2):219–224. doi:10.2108/zsj.23.219.
- Suk HY, Jeon JY, Kim D-Y, Cha S, Min M-S. 2021. The complete mitochondrial genome information of Rana uenoi (Amphibia, Anura, Ranidae) and the phylogenetic implication. Mitochondrial DNA B Resour. 6(2):689–690. doi:10.1080/23802359.2021.1882896.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi:10.1093/molbev/msab120.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One. 9(11):e112963. doi:10.1371/journal.pone.0112963.
- Wang J, Li Z, Gao H, Liu Z, Teng L. 2020. The complete mitochondrial genome of the Rana kukunoris (Anura: Ranidae) from Inner Mongolia, China. Mitochondrial DNA B Resour. 5(1):586–587. doi:10.1080/23802359.2019.1710591.
- Yang BT, Zhou Y, Min MS, Matsui M, Dong BJ, Li PP, Fong JJ. 2017. Diversity and phylogeography of Northeast Asian brown frogs allied to Rana dybowskii (Anura, Ranidae). Mol Phylogenet Evol. 112:148–157. doi:10.1016/j.ympev.2017.04.026.
- Zhou Y, Yang B-T, Li P-P, Min M-S, Fong JJ, Dong B-J, Zhou Z-Y, Lu Y-Y. 2015. Molecular and morphological evidence for Rana kunyuensis as a junior synonym of Rana coreana (Anura: Ranidae). J Herpetol. 49(2):302–307. doi:10.1670/13-111.
