Abstract
Duck breed Longshengcui (Anas platyrhynchos Linnaeus, 1758 breed Longshengcui, LSC) is one of the famous native breed of the Guangxi Zhuang Nationality Autonomous Region in China. In this study, we report the complete mitochondrial genome of LSC. The mitogenome (GenBank accession no. MZ895120) has 16,602 bp in length and consisted of the well-known 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and the control region. The phylogenetic analysis showed that LSC and Zhijiang duck have highly similar genetic relationship. These results are helpful for the conservation of genetic resources and phylogeny of this species.
Duck breed Longshengcui (Anas platyrhynchos Linnaeus, 1758 breed Longshengcui, LSC) is one of the famous native breeds of the Guangxi Zhuang Nationality Autonomous Region in China (). LSC is a rare genetic resource of duck species with its high ornamental quality and tender meat. It is important to preserve the genetic resources of native species for urgent conservation of the species. A complete mitochondrial genome is one of the unique genetic characters suitable for the purpose and plays an important role in phylogenetic relationships discovery (Zhai et al. Citation2021). In this study, we report LSC’s complete mitochondrial genome sequence.
The adult individuals of LSC were collected at its originally breeding farm in Longsheng County (25°29′N and 109°43′28″E), Guilin City, Guangxi Zhuang Nationality Autonomous Region. And the specimens were stored at −80 °C in our laboratory (School of life sciences Guangxi Normal University, Guilin, http://www.bio.gxnu.edu.cn/, Xin Wang is the contact person [email protected]) under the voucher number GXNU-LSC-202105066. Total genomic DNA was extracted from the thorax muscle of a single individual using the EasyPure Kit of Genomic DNA (Transgen Biotech, Beijing, China). Polymerase chain reaction (PCR) was carried out to amplify the complete mitochondrial genome with 13 pairs of primers. PCR products of the gel electrophoresis were purified by Gel AdvancedTM Gel Extraction (Rich Biotech, Taiwan, China) and sequenced by BioSune Biotech (Shanghai, China) and using an ABI 3730 automatic sequencer (Sanger sequencing). The complete mitochondrial genome sequence was assembled manually using DNAstar v7.1 software. The LSC duck mitogenome was annotated using DOGMA (Wyman et al. Citation2004).
The complete circular mitogenome of LSC presented 16,602 bp in size (GenBank accession no. MZ895120). Its mitogenome contains 13 protein-coding genes which were identified by an ORF finder analysis at NCBI (https://www.ncbi.nlm.nih.gov/orffinder/), and two ribosomal RNA genes were identified by sequence alignments and compared to several related species. Moreover, 22 transfer RNA genes were identified with tRNAscan-SE (Chan and Lowe Citation2019); and a control region (D-loop) of 1048 bp was also identified. The 12S rRNA and 16S rRNA lengths were 985 and 1602 bp. The OGDRAW version 1.3.1 was used to draw the physical map of the complete genome () (Greiner et al. Citation2019). Most of the genes were located on the heavy chain, except for ND6 and eight tRNAs genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, tRNAGlu), which were similar to most birds mtDNAs (Lin et al. Citation2016, Lin et al. Citation2016, Liu et al. Citation2021). For 13 PCGs in the LSC mtDNA, besides the COX1, COX2, ND5 initiation codon is GTG, ND6 is CTA, and the rest of the PCGs are ATG. LSC mt DNA had five types of termination codons, including AGG (ND1 and COX1), TAG (ND2), TAA (COX2, ATPase8, ATPase6, ND3, ND4L, ND5 and Cytb), CAT (ND6) and an incomplete termination codon ‘‘T–’’ (COX3 and ND4). LSC duck mtDNA possess TAA or TAG as their termination codon and this difference in termination codon is also frequently found in other mitogenomes () (Jia et al. Citation2023).
Figure 2. Mitochondrial genome map of Longshengcui duck. The inner circle indicates the GC content, and the external circle indicates the genes having different colors based on their functions. The arrows represent direction of transcription, genes encoded on the heavy and light strand are shown outside and inside the circle, respectively.
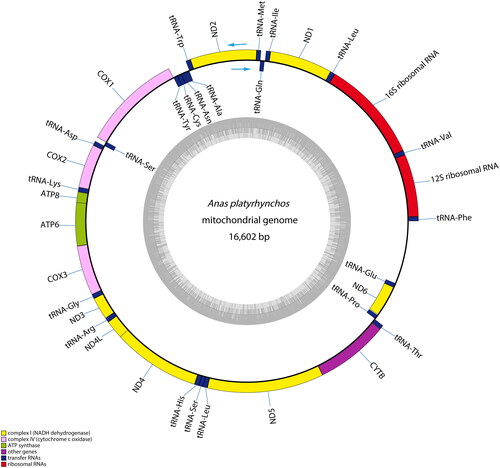
Table 1. Organization of the mitochondrial genome of Longshengcui duck.
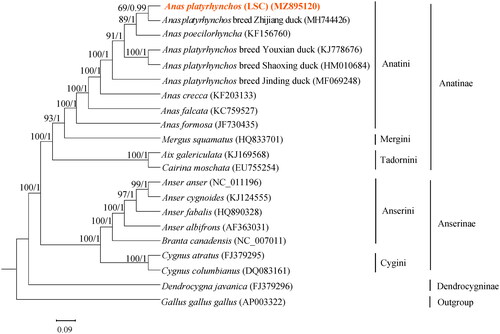
Phylogenetic analysis was performed using the complete mitochondrial DNA sequences of 20 Anseriforms and outgroup mitogenomes. Each of the sequences datasets was aligned by ClustalX (Thompson et al. Citation1997). Phylogenetic tree was inferred by Maximum Likelihood (ML) and Bayesian Inference (BI) models and constructed via IQtree (Nguyen et al. Citation2015) and MrBayes 3.2.7 (Huelsenbeck and Ronquist Citation2001), respectively. The ML/BI tree showed that the analyzed species are divided into three major clades: Anatinae, Anserinae and Dendrocygninae (). Anatinae is the first lineage, which is sister to the second group, Anserinae; Dendrocygninae forms the third group and is sister to Anatinae and Anserinae. The outgroup, Gallus gallus gallus, is located at the base of the tree. The first lineage Anatinae includes tribes Anatini (Anas platyrhynchos breed LSC, Anas platyrhynchos breed Zhijiang duck, Anas poecilorhyncha, Anas platyrhynchos breed Youxian duck, Anas platyrhynchos breed Shaoxing duck, Anas platyrhynchos breed Jinding duck, Anas crecca, Anas falcata, Anas formosa), Mergini (Mergus squamatus), Tadornini (Aix galericulata, Cairina moschata). The second lineage includes tribes Anserini (Anser anser, Anser cygnoides, Anser fabalis, Anser albifrons and Branta canadensis) and Cygnini (Cygnus atratus and Cygnus columbianus), forming the subfamily Anserinae. The third lineage Dendrocygninae groups together with Anatinae and Anserinae. Meanwhile, we also found that LSC and Anas platyrhynchos breed Zhijiang duck having a highly similar genetic composition.
Figure 3. Phylogenetic analysis based on complete mitochondrial genome sequences. An ML/BI tree was built based on the phylogenetic analysis of 20 Anseriform species’ complete mitochondrial genomes. The mitochondrial genome sequences of the Anseriform species were obtained from the GenBank databases (accession numbers have marked on the figure). Abbreviation of species indicates: LSC, Longshengcui duck.
Ethics statement
All the experiment procedures were approved by the Animal Care Committee of School of life sciences Guangxi Normal University, Guilin, China.
Authors’ contributions
The research was designed and conducted by QL, CJL, SYB. The animal experiment was conducted by JMZ, XW and QXW. The detection and analysis works were conducted by JMZ, XW, QXW, LPL, FMC and YZW. The manuscript draft was prepared by QL, CJL, SYB, JMZ, XW and QXW. All authors participated in the discussion and editing of the manuscript.
Supplemental Material
Download MS Word (22.2 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ895120.
Additional information
Funding
References
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. doi: 10.1093/bioinformatics/17.8.754.
- Jia Y, Qiu G, Cao C, Wang X, Jiang L, Zhang T, Geng Z, Jin S. 2023. Mitochondrial genome and phylogenetic analysis of Chaohu duck. Gene. 851:147018. doi: 10.1016/j.gene.2022.147018.
- Lin Q, Jiang G-T, Yun L, Li G-J, Dai Q-Z, Zhang S-R, Hou D-X, He X. 2016. The complete mitochondrial genome of the Linwu duck. Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):992–993. doi: 10.3109/19401736.2014.926520.
- Lin Q, Qiu L, Cao R, Jiang G-T, Dai Q-Z, Zhang S-R, Hou D-X, He X. 2016. The complete mitochondrial genome of the Youxian duck. Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):990–991. doi: 10.3109/19401736.2014.926519.
- Liu D, Zhou Y, Fei Y, Xie C, Hou S. 2021. Mitochondrial genome of the critically endangered Baer’s Pochard, Aythya baeri, and its phylogenetic relationship with other Anatidae species. Sci Rep. 11(1):24302. doi: 10.1038/s41598-021-03868-7.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882. doi: 10.1093/nar/25.24.4876.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. doi: 10.1093/bioinformatics/bth352.
- Zhai H, Li ZZ, Mi SH, Meng DH, Yu HX, Teng LW, Liu ZS. 2021. The complete mitochondrial genome of the Ferruginous Duck (Aythya nyroca) from Ningxia, China. Mitochondrial DNA B Resour. 6(2):546–547. doi: 10.1080/23802359.2020.1870901.

