Abstract
Aster altaicus Willd. is an important medicinal plant and can also be used as a forage grass. To better understand the diversity and phylogeny between A. altaicus and other Aster species, we sequenced and annotated the complete chloroplast genome of A. altaicus by using the Illumina Hiseq 2500 platform. This complete chloroplast genome is 152,473 bp long and the GC content is 37.3% presented a negative AT-skew (–0.002) and a positive GC-skew (0.003). The genome contains a large single-copy region (LSC) of 84,235 bp, a small single-copy region (SSC) of 18,212 bp, which separated by a pair of inverted repeat regions (IRA and IRB) of 25,013 bp. Moreover, 129 genes were found in the chloroplast of A. altaicus, including 85 protein-coding genes (PCGs), 36 transfer RNA genes (tRNAs), 8 ribosomal RNA unit genes (rRNAs). Phylogenetic analysis showed that A. altaicus was more closely related to A. altaicus and A. altaicus var. uchiyamae. This study lays the foundation for further studies on the evolution and phylogeny of Aster.
Introduction
Aster altaicus Willd. is a perennial herb () (Willdenow Citation1809) that can grow in grasslands, deserts, sands, and arid mountainous areas at altitudes of 0–4000 m. It has a large distribution in northern China and also grows in other Asian countries. As an important medicinal plant, it is widely used in TCM (traditional Chinese medicine) and Mongolian medicine (Ma and Fu Citation2000), and possesses some forage value (Jia Citation1987). In the early stage of growth, goats like to eat its fresh branches and leaves, and sheep like to eat its branches and flowers. Since the complete chloroplast genome sequence of A. altaicus has not been reported and its genetic composition and phylogenetic relationships remain unclear. Thus, leaf tissue for this study was extracted by using the TruSeq DNA Sample Vegetation Kit (Vanzyme, China) for total genomic DNA, followed by sequencing using the Illumina Hiseq 2500 platform (Illumina, SanDiego, CA), with 150 bp paired-end reads provided by Genesky Biotechnologies Inc, Shanghai, China. After trimming, the high-quality paired-end reads were assembled through metaSPAdes software (version 3.13.0) (Korobeynikov Citation2017). The present study reports the complete chloroplast (cp) genome of A. altaicus collected from the Qinghai-Tibetan plateau for the first time, which helps provide more information about the plastid evolution and intrageneric diversity of Aster. It has important implications for further understanding the evolution and phylogeny of Aster.
Materials and methods
The fresh leaves of a single individual were collected from Yueliang Bay Park in Guide County, Hainan Tibetan Autonomous Prefecture, Qinghai Province, China (101°43’95″ E, 36°04′61″N), in August 2022. After on-site collection, the samples were immediately flash-frozen in liquid nitrogen and then directly mailed to the sequencing company (Biotechnologies Inc). The specimens (voucher no. QHHNYLW22-B3) have been deposited in the Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau, College of Qinghai Academy of Animal and Veterinary Sciences, Qinghai University, Xining, China (URL: https://mky.qhu.edu.cn/index.htm, contact person: Ying Liu and email is [email protected]).
The total genomic DNA was extracted from a single specimen using a DNeasy Tissue Kit (Qiagen, Germany). The total read length of raw data was 22,184,764 bp. The following information describes how the complete plastome was assembled. The sequence was matched to the genome, and all positions read mapping depth of assembled genome was statistically obtained, respectively (Figure S1). In the process, the complete chloroplast genome of A. flaccidus (GenBank accession NC 052918) was used as a reference genome, and the genome annotation was performed with the program by CPGAVAS2 software (version N/A) (Shi et al. Citation2019) comparing the sequences with the complete chloroplast genome of A. flaccidus. Then, we corrected annotations by comparing them with the published complete chloroplast genomes of Aster species. The map of cis/trans-splicing gene and plastome using the CPGView program (http://www.1kmpg.cn/cpgview) (Liu et al. Citation2023). Finally, the assembled chloroplast genome and its detailed annotations were submitted to GenBank under the accession number NC072176.
Phylogenetic analysis was performed through the following procedure: A total of 25 Aster chloroplast genomes were downloaded from GenBank, Medicago monspeliaca (MK460506), and Medicago radiata (MK460505) were used as outgroups, and were aligned using MEGA7(Kumar et al. Citation2016) with default parameter. The maximum likelihood (ML) tree was built using MEGA7 with bootstrap set to 1000.
Results
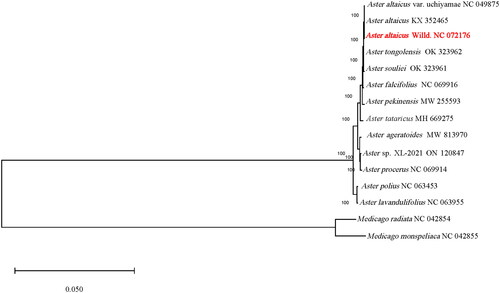
We obtained the complete chloroplast genome of A. altaicus is a circular DNA molecule (), with 152,473 bp long, a GC content is 37.3% and presented a negative AT-skew (−0.002), and a positive GC-skew (0.003) on the J-strand. The genome contains one large single-copy region (LSC) with 84,235 bp, one small single-copy region (SSC) with 18,212 bp and two inverted repeat regions (IR) with 25,013 bp. This complete chloroplast genome encodes a total of 129 genes, including 85 protein-coding genes (PCGs), 36 transfer RNA genes (tRNAs), 8 ribosomal RNA unit genes (rRNAs). Among these genes, 17 genes (rps16, rpoC1, atpF, petB, petD, rpl16, rpl12, ndhB, ndhA, rpl2, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, and trnI-GAU) have one intron each, and 3 genes (ycf3, rps12, and clpP) contain two introns each. A total of 70 SSR markers ranging from mononucleotide to pentanucleotide repeat motif were identified in the A. altaicus chloroplast genome. Phylogenetic analysis of the complete chloroplast sequences of A. altaicus and 12 species of the genus Aster in the family Asteraceae. Two Medicago species (Medicago monspeliaca and Medicago monspeliaca) of Fabaceae were used as outgroups. Phylogenetic analysis indicated a strong sister relationship with A. altaicus and Aster altaicus var. uchiyamae (). The complete chloroplast genome of A. altaicus will contribute to a better understanding of the evolutionary mode of the chloroplast genome and provide more evidence for the identification and application of Aster species.
Figure 2. The plastome genome map of Aster altaicus under this study. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). the small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
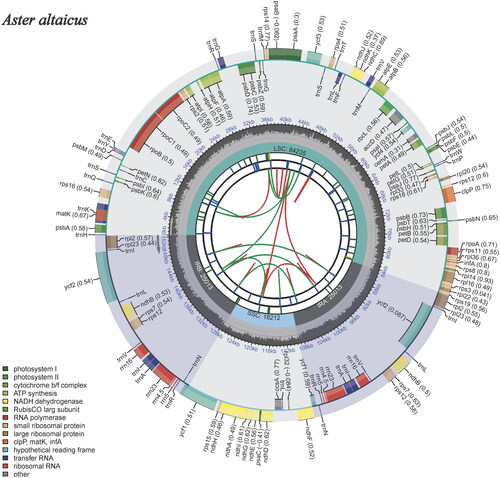
Figure 3. Chloroplast phylogeny of 13 Aster species based on the complete chloroplast genome sequences. The red fonts represents the assembled plastome sequence in this study. The clades of species are represented with black lines. The following sequences of each species were used: Aster ageratoides MW 813970 (Feng et al. Citation2021), Aster altaicus var. uchiyamae NC 049875, Aster altaicus KX 352465 (Park et al. Citation2017), Aster altaicus NC 072176 (this study), Aster falcifolius NC 069916, Aster lavandulifolius NC 063955, Aster pekinensis MW 255593 (Zhang et al. Citation2021), Aster polius NC 063453, Aster procerus NC 069914, Aster souliei OK 323961 (Wang et al. Citation2022), Aster sp. XL-2021 on 120847, Aster tataricus MH 669275 (Shen et al. Citation2018), Aster tongolensis OK 323962(Wang et al. Citation2022), Medicago monspeliaca NC 042855(Choi et al. Citation2019), Medicago radiata NC 042854 (Choi et al. Citation2019). two Medicago species (Medicago radiata and Medicago monspeliaca) of Fabaceae were used as outgroups. Undescribed citations in the legend indicate that the citations have not been published.
Discussion and conclusion
A. altaicus Willd. is a plant of Aster in the Asteraceae family. The genus of Aster is a relatively large genus in the Asteraceae family, wide distributed in China (Lin et al. Citation1985). It has been the focus of phylogenetic studies. In this research, we successfully assembled and annotated the complete chloroplast genome of A. altaicus Willd. collected from the Qinghai-Tibetan plateau for the first time. With a sequence length of 152,473 bp, the entire chloroplast genome has a sequence length similar to that of other Aster sp. It also has the typical quadripartite structure of the chloroplast genome of most angiosperms. The taxonomic affiliation of the genus of Aster is currently controversial. Some studies suggesting that some species should not be included in the Aster, such as Aster albescens, Aster argyropholis and Aster asteroids et al. (Li et al. Citation2012). To avoid ambiguity, the phylogenetic analysis in this study didn’t include the chloroplast genome sequences of these controversial species. Notably, the phylogenetic analysis showed that A. altaicus Willd. is more closely related to A. altaicus and A. altaicus var. uchiyamae collected in Korea. This study provides important genetic resource information for further studies of A. altaicus Willd. It is important to study the phylogenetic relationships of Aster. In the future, we should continue to strengthen the genetic information mining of Aster sp. and further improve the taxonomic study of Aster sp. and even Asteraceae.
Ethical approval
Based on No. 221(E) Article 15: (1)-(ii) of the International Union for the Protection of New Varieties of Plants (UPOV) in 1991, this study can be conducted without ethical approval or permission. And research on plant chloroplast genome sequencing does not require ethical approval.
Author contributions
Ying Liu (corresponding author) collected the materials, conception and designed the experiment. Xin-You Wang (first author) analyzed and interpretation of the data, and drafting of the manuscript. After that, Ying Liu revised it critically for intellectual content of this article and agreed to the final approval to be published. All authors reviewed the manuscript and agreed to be accountable for all aspects of the work.
Plant material collection statement
The species is widely distributed in China; the plant materials were not obtained from nature reserves, and the chloroplast genome sequencing research did not affect the population, so collection licenses were not needed.
Supplemental Material
Download TIFF Image (394.4 KB)Supplemental Material
Download TIFF Image (247 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank database of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession No. NC072176. The associated BioProject, and BioSample numbers are PRJNA905941 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA905941), SRA: SRR22438481 (https://www.ncbi.nlm.nih.gov/sra/?term= SRR22438481), and SAMN31888589 (https://www.ncbi.nlm.nih.gov/biosample/?term= SAMN31888589), respectively.
Additional information
Funding
References
- Choi IS, Jansen R, Ruhlman T. 2019. Lost and found: return of the inverted repeat in the legume clade defined by its absence. Genome Biol Evol. 11(4):1321–1333.
- Feng J-Y, Wu Y-Z, Wang R-R, Xiao X-F, Wang R-H, Qi Z-C, Yan X-L., 2021. The complete chloroplast genome of balsam aster (Aster ageratoides Turcz. var. scaberulus (Miq.) Ling., Asteraceae). Mitochondrial DNA B Resour. 6(9):2464–2465. doi: 10.1080/23802359.2021.1955030.
- Jia SX. 1987. Forage plants of China. Vol. 1. Beijing: Agricultural Press.
- Korobeynikov A. 2017. SPAdes: is there anything new we could develop? http://programme.exordo.com/sfaf2017/delegates/presentation/92/
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Li WP, Yang FS, Jivkova T, Yin GS. 2012. Phylogenetic relationships and generic delimitation of Eurasian Aster (Asteraceae: Astereae) inferred from ITS, ETS and trnL-F sequence data. Ann Bot. 109(7):1341–1357. doi: 10.1093/aob/mcs054.
- Lin R, Chen YL, Shi Z. 1985. Flora of China. Vol.74. Beijing: Science Press.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Ma YQ, Fu XQ. 2000. Flora intramongolica. Vol. 4. Huhhot: Innermongolian People’s Publsihing House. P. 495.
- Park J, Shim J, Won H, Lee J. 2017. Plastid genome of Aster altaicus var. uchiyamae Kitam., an endanger species of Korean asterids. Journal of Species Research. 6(1):76–90.
- Shen X, Guo S, Yin Y, Zhang J, Yin X, Liang C, Wang Z, Huang B, Liu Y, Xiao S. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules. 23(10):1–9. doi: 10.3390/molecules23102426.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nuclc Acids Research. 47:65–73.
- Wang J, Zhao R, Su X, Amu H, Zhang Z. 2022. The complete chloroplast genome sequences of Aster souliei Franch and Aster tongolensis Franch (Asteraceae). Mitochondrial DNA B Resour. 7(3):523–524. doi: 10.1080/23802359.2022.2054375.
- Willdenow DCL. 1809. Enumeratio horti regii botanici berolinensis. Berolini: Taberna Libraria Scholae Realis.
- Zhang X, Jiang PP, Fan SJ. 2021. Characterization of the complete plastome of Aster pekinensis (Asteraceae), a perennial herb. Mitochondrial DNA B Resour. 6(3):1064–1065. doi: 10.1080/23802359.2021.1899081.

