Abstract
Aegle marmelos (L.) Correa 1800, a plant belonging to the Rutaceae family, is extensively used in Tibetan medicine. We employed Illumina HiSeq reads to assemble the complete chloroplast (cp) genome of A. marmelos, which spans 144,538 bp. The genome comprises 114 genes, including 75 protein-coding genes, 31 tRNA genes, and 8 rRNA genes. It is characterized by four regions: The large single-copy (LSC) region (74,253 bp), the inverted repeat A (IRa) region (26,015 bp), the small single-copy (SSC) region (18,255 bp), and the inverted repeat B (IRb) region (26,015 bp). Phylogenomic analysis demonstrated a close relationship between A. marmelos and Citrus. The assembly of The cp genome in this study serves as a foundation for conservation efforts and phylogenetic investigations of A. marmelos, paving the way for future experimentation.
Introduction
Aegle marmelos (L.) Correa, a 10-m-tall tree of the Rutaceae family (), holds significant importance in Tibetan medicine. Our team captured local images of this species with a professional team. While It is indigenous to India, it is also found in Southeast Asia and has been introduced to Florida and Hawaii (Maity et al. Citation2009). A. marmelos is native to Xishuangbanna in Yunnan Province, as well as India, Burma, Laos, Vietnam, Cambodia, Thailand, Malaysia, and Indonesia, where it thrives in slightly dry, sloping forests at altitudes of 600–1000 m. All parts of the plant, including the roots, bark, leaves, flowers, fruits, seeds, and latex, are utilized in traditional medicinal systems (Patel et al. Citation2012). The plant possesses numerous pharmacological properties, with Its roots, bark, leaves, and flowers being employed in the treatment of intermittent fevers. The fleshy part of the fruit serves as a gastrointestinal medicine and dietary supplement. In Myanmar, young leaves are utilized to treat wounds, scabies, swelling, pain, and foot-and-mouth disease. Additionally, the juice derived from crushed leaves is employed to treat eye diseases. However, caution must be exercised by Pregnant women and those preparing for pregnancy, as the consumption of young industry from this species can lead to infertility and even miscarriage (Rana et al. Citation1997; Charoensiddhi and Anprung Citation2008). This review focuses on the investigation of the chloroplast (cp) genome of A. marmelos and its phylogenetic position within the Rutaceae family. We assembled and characterized the complete cp genome of A. marmelos, enhancing our understanding of its genetic information and facilitating species identification within the Rutaceae family.
Figure 1. Images of Aegle marmelos (L.) Correa 1800: (a) Leaves; (b) fruit; (c) whole plant. A. marmelos is a deciduous small tree, approximately 10 m tall, with leaflets featuring numerous oil glandular dots. The leaflets are broadly ovate or long elliptic, measuring 4–12 cm in length. terminal leaflets are larger and long-stalked, while lateral leaflets are subsessile with shallowly obtuse serrations. The fruit measures 10–12 cm in height and 6–8 cm in diameter, displaying a light greenish-yellow color, smooth texture, and a hard woody pericarp, approximately 3–4 mm thick. The fruit stalk is 4–6 cm long, and the seeds, numerous in quantity, are compressed ovoid with transparent gum. The testa is wooly, and cotyledons emerge during germination. These photos were captured by a local professional team in Xishuangbanna County, Yunnan Province, China.

Materials and methods
Plant material and DNA sequencing
This study was conducted in compliance with the Regulations on Biodiversity Conservation of Yunnan Province and received authorization from Xishuangbanna (Yunnan Province, China) and the Institute of Chinese Materia Medica, Chinese Academy of Chinese Medical Sciences (Beijing, China). permission was obtained from the local forestry department for sample collection. Kunming Caizhi Biotechnology Co., Ltd. (Kunming, China) identified the A. marmelos specimen. A. marmelos impatiens samples were obtained from Mengla County, Xishuangbanna Prefecture, Yunnan Province, China (alt. 2683 m, E101°15′12″, N21°55′40″). The voucher specimen (specimen accession number: LQE-2022-0523) was deposited in the Institute of Chinese Meteria Medica, China Academy of Chinese Medical Sciences (contact: Xiangxiao Meng, [email protected]). Total DNA was extracted from silica-dried young leaves using a Plant genomic DNA kit (Tiangen Biotechnology (Beijing) Co., Ltd., Beijing, China).
Genome assembly and annotation
In this study, the chloroplast genome was annotated using GetOrganelle v.1.7.5 (Jin et al. Citation2020) and CPGAVAS2 (http://www.herbalgenomics.org/cpgavas2). the complete chloroplast genome assembly was performed using Illumina HiSeq reads, and the C. inopinata chloroplast genome (Genome Repository accession number: Gwhbk01000000) was confirmed through a BLAST search.
Phylogenetic analysis
A maximum-likelihood (ML) method was employed to reconstruct the phylogenetic relationships between A. marmelos and 14 other species. The chloroplast genomes were downloaded from the NCBI GenBank database, with Ruta graveolens (NC_045946) serving as an outgroup.
The cp genomes were aligned using MAFFT v. 7.471 (Katoh and Standley Citation2013) and trimmed. A maximum likelihood phylogenetic tree was constructed using RAxML v8.2.12 (Alexandros Citation2014), employing the GTRGAMMA model, 1,000 bootstrap replicates, and the rapid bootstrapping and search for best-scoring ML tree algorithm. The resulting phylogenetic tree () was modified using iTOL (Ivica and Bork Citation2019).
Result and discussion
Genome structure analysis
To ensure high-quality reads, the raw sequencing data (raw reads) from the offline machine were filtered to remove low-quality reads with adapters, resulting in clean reads and ensuring the accuracy of subsequent analyses. The genome assembly achieved 100% coverage (Figure S1).
DNA fragmentation was performed using an ultrasonic processor, yielding DNA fragments with an insert length of approximately 144,538 bp. The library preparation involved processes such as terminal repair, addition of base A, addition of sequence adapters, purification, and PCR amplification to obtain a 144,538 bp library. Novenol Co., Ltd. (Tianjin, China) and the Novena vaSeq System (Novena, Santiago, CA) performed whole-genome sequencing. The chloroplast genome was assembled using SPAdes version 3.13.0 with default settings, without employing a reference genome.
The complete chloroplast genome of A. marmelos (Genome Warehouse accession number: Gwhbkhk01000000) spans a length of 144,538 bp, with a GC content of 38.9%. It consists of four regions: two inverted repeat regions (IRa and IRb) of 26,015 bp each, separated by a large single-copy gene region (LSC) of 74,253 bp, and a small single-copy gene region (SSC) of 18,255 bp. The chloroplast genome contains a total of 114 genes, including 75 protein-coding genes, 31 tRNA genes, and 8 rRNA genes (). Among these genes, nine were found to contain introns. Seven genes (rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) contain a single intron, while two genes (ycf3 and clpP) contain two introns (Figure S2).
Figure 2. Chloroplast reference genome of Aegle marmelos. The diagram illustrates the organization of the chloroplast genome. Starting from the center, the first circle represents the forward and reverse repeat sequences, indicated by red and green arcs, respectively. The second circle depicts a tandem repeat sequence, while the third circle highlights microsatellite sequences with short strips. The fourth circle displays the positions of the large single-copy gene region (LSC), small single-copy gene region (SSC), and inverted repeat regions (IRA and IRB). the fifth circle represents the GC content. The genes located outside the sixth circle are transcribed counterclockwise, while those inside are transcribed clockwise. Different functional groups of genes are color-coded, as indicated in the legend.
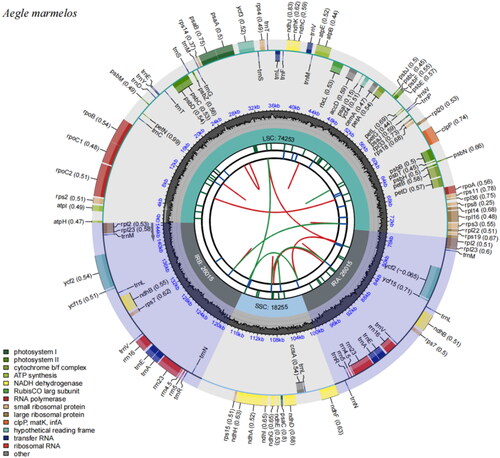
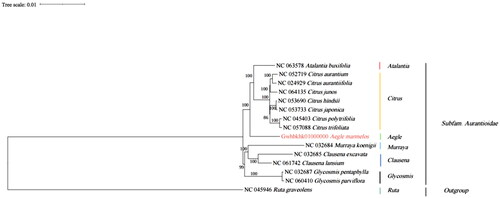
Figure 3. Showcases the phylogenetic analysis of 14 species and 1 taxonomic unit outgroup, utilizing the complete chloroplast genome sequence with the RAxML method. The following sequences were used for analysis: NC_063578 Atalantia buxifolia (Wang et al. Citation2017), NC_052719 Citrus aurantium (Lin et al. Citation2020), NC_024929 Citrus aurantiifolia (Su et al. Citation2014), NC_064135 Citrus juno (Zhong-Wang Citation2008), NC_053690 Citrus hindsii (Xu et al. Citation2019), NC_053733 Citrus japonica (Wu et al. Citation2019), NC_045403 Citrus polytrifolia (Li et al. Citation2019), NC_057088 Citrus trifoliata, Gwhbkhk01000000 Aegle marmelos, NC_032684 Murraya koenigii (Shivakumar et al. Citation2017), NC_032685 Clausena excavata (Natthawadi et al. Citation2018), NC_061742 Clausena lansium (Zhao et al. Citation2019), NC_032687 Glycosmis pentaphylla (Shivakumar et al. Citation2017), NC_060410 Glycosmis parviflora (Wen-Fang et al. Citation2013), and NC_045946 Ruta graveolens (Ling and Zhang Citation2019).
Phylogenetic analysis
Aegle marmelos belongs to the genus Aegle, and no similar structure is found within the genus. Therefore, a phylogenetic tree was constructed using 14 species from different genera within the same subfamily, along with one outgroup species from a different subfamily. The complete chloroplast genome sequence of A. marmelos provides valuable information for species conservation and phylogenetic studies of the Rutaceae family.
Authors’ contributions
Sijia Wang completed the article. Rushang Xiang uploaded the genomic data, using the database of the Chinese Academy of Traditional Chinese Medicine, Xiubo Liu and Jiaxin Lu analyzed the data in the article, Xiubo Liu created the graphs in the article, and Lingyang Kong and Zhanping Zhang conducted some literature searches. Wei Ma designed the experiments. All authors agreed to take responsibility for all aspects of the work.
Endangered species statement
This article adheres to the International Union for Conservation of Nature (IUCN) policy research involving endangered species and strictly adheres to the Convention on Biological Diversity and the Convention on Trade in Endangered Species of Wild Fauna and Flora (CITES) without damaging endangered species.
Supplemental Material
Download MS Word (197 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data obtained in this study are openly available in GenBank of NGDC (https://ngdc.cncb.ac.cn/) under the accession number Gwhbkhk01000000. The associated BioProject, Bio-Sample and GSA numbers are PRJCA011521, SAMC866117 and CRA007998, respectively.
Additional information
Funding
References
- Alexandros, SJ. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.
- Charoensiddhi S, Anprung PJ. 2008. Bioactive compounds and volatile compounds of Thai Bael Fruit (Aegle Marmelos (L.) Correa) as a valuable source for functional food ingredients.
- Ivica L, Bork P. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. (W1). W256–W259.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 1(21):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li S, Zong D, Zhou A, He C. 2019. The complete chloroplast genome sequence of Poncirus polyandra (Rutaceae), an endangered species endemic to Yunnan Province, China. Mitochondrial DNA Part B. 4(1):766–768. doi: 10.1080/23802359.2019.1565974.
- Lin H, Li X, Bai D. 2020. Assembly and phylogenetic analysis of the complete chloroplast genome of Citrus aurantium (Rutaceae). Mitochondrial DNA B Resour. 5(3):2250–2251. doi: 10.1080/23802359.2020.1771228.
- Ling LZ, Zhang SD. 2019. The complete chloroplast genome of the multipurpose and traditional herb, Ruta graveolens L. Mitochondrial DNA B Resour. 4(2):3661–3662. doi: 10.1080/23802359.2019.1678426.
- Maity P, Dhananjay B, Uday M, Kumar DJ. 2009. Biological activities of crude extracts and chemical constituents of Bael. Aegle Marmelos (L.) Corr. 47(11):849–861.
- Natthawadi W, Nuttha S, Johann S, Karin VV, Markus B, Srunya, VJBS, Ecology. 2018. Chemodiversity of Clausena excavata (Rutaceae) and related species. Coumarins Carbazoles. 80:84–90.
- Patel PK, Sahu J, Sahu L, Prajapati NJ. 2012. Aegle marmelos: a review on its medicinal properties.
- Rana BK, Singh UP, Taneja V. 1997. Antifungal activity and kinetics of inhibition by essential oil isolated from leaves of Aegle marmelos. J Ethnopharmacol. 57(1):29–34. doi: 10.1016/s0378-8741(97)00044-5.
- Shivakumar VS, Appelhans MS, Johnson G, Carlsen M, Zimmer EA. 2017. Analysis of whole chloroplast genomes from the genera of the Clauseneae, the Curry tribe (Rutaceae, Citrus family). Mol Phylogenet Evol. 117:135–140. doi: 10.1016/j.ympev.2016.12.015.
- Su HJ, Hogenhout SA, Al-Sadi AM, Kuo CH. 2014. Complete chloroplast genome sequence of omani lime (Citrus aurantiifolia) and comparative analysis within the Rosids. 9.
- Wang X, Xu Y, Zhang S, Cao L, Huang Y, Cheng J, Wu G, Tian S, Chen C, Liu Y, et al. 2017. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet. 49(5):765–772. doi: 10.1038/ng.3839.
- Wen-Fang MA, Zhu XY, Cai Y, Zhao XZ, Jiang JP. 2013. Extraction of the volatile compounds from Folium glycosmis parviflorae by steam distillation and supercritical fluid extraction.
- Wu JW, Liu F, Tian N, Liu JP, Cheng CZ. 2019. Characterisation of the complete chloroplast genome of Fortunella Crassifolia Swingle and phylogenetic relationships. Mitochondrial DNA B Resour. 4(2):3538–3539.
- Xu SR, Zhang YY, Liu F, Tian N, Pan DM, Bei XJ, Cheng CZ. 2019. Characterization of the complete chloroplast genome of the Hongkong kumquat (Fortunella hindsii Swingle). Mitochondrial DNA B Resour. 4(2):2612–2613. doi: 10.1080/23802359.2019.1642166.
- Zhao ZC, Gao AP, Huang J. 2019. Sequencing and analysis of chloroplast genome of Clausena lansium (Lour.) Skeels.
- Zhong-Wang L. 2008. Characterization of malate dehydrogenase gene from Citrus junos and its transgenic tobacco’s tolerance to aluminium toxicity.
