Abstract
Carallia brachiata (Lour.) Merr. (1919) is an important medical resource distributed across subtropical Asia. In this study, the complete chloroplast genome of C. brachiata was sequenced, revealing a total length of 162,460 bp, including four regions – a large single copy (89,814 bp), a small single copy (18,804 bp), and a pair of inverted repeats (26,921 bp each). The overall guanine + cytosine content was 35.76%. In total, 130 genes were annotated within the chloroplast genome, comprising 85 protein-coding, 37 tRNA, and 8 rRNA genes. Subsequent phylogenetic analyses revealed that C. brachiata is closely related to Carallia diplopetala.
Introduction
Carallia brachiata (Lour.) Merr. (1919) is a member of the genus Carallia in the family Rhizophoraceae and is mostly distributed across subtropical Asia. Its leaves are oval and have smooth surfaces and edges (). It is an important medical resource for treating sapraemia, and its bark is used in pruritis treatment (Ling et al. Citation2004). However, there is no record of the complete chloroplast genome of C. brachiata in the National Center for Biotechnology Information (NCBI) database. Therefore, in this study, the complete chloroplast genome of C. brachiata was sequenced, and its phylogenetic position within the Rhizophoraceae was confirmed.
Materials and methods
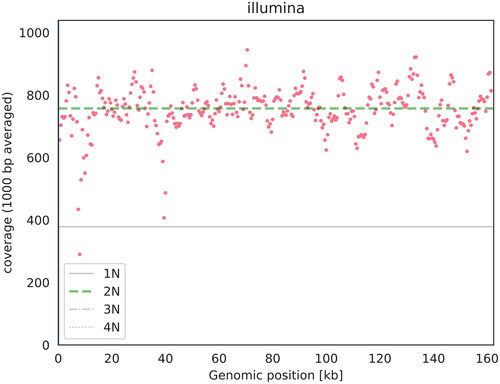
Fresh leaves of C. brachiata were collected from the South China Botanical Garden in Guangzhou, China (23°11’16.8" N, 113°22’15.6″ E) in compliance with the national Wild Plant Protective Regulations. The Biomarker Technologies Corporation (Beijing, China) approved the collection of the required samples for analysis. A specimen was deposited at Biomarker Technologies Corporation (Jian Zhao, email: [email protected]) under the voucher number ZJS202101110ZJ. The total genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987), and libraries were prepared using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA). The libraries were then sequenced on the Illumina Novaseq 6000 platform, and the raw data obtained were filtered using PRINSEQlite v. 0.20.4 (Schmieder and Edwards Citation2011), yielding 3.46 Gb of clean data with a read coverage depth over 600X (). High-quality reads were assembled into the chloroplast genome using de novo assembler SPAdes v.3.11.0 (Bankevich et al. Citation2012). Finally, the complete chloroplast genome was annotated using the PGA software package (Qu et al. Citation2019), with the chloroplast genome of Pellacalyx yunnanensis (MN106253) serving as a reference. The results were then submitted to GenBank under accession number OM141003.
Sixty-two homologous protein-coding genes (PCGs) from 26 chloroplast genomes in the NCBI were selected using OrthoFinder v2.3.14 (Emms and Kelly Citation2015). These were aligned with the C. brachiata genome using MUSCLE v.3.8.1551 (Edgar Citation2004), and conserved sequences were extracted from the alignment using Gblocks v0.91b (Talavera and Castresana Citation2007). Prottest v3.4 was used to select the HIVb + I + G + F model, and Couratari macrosperma (MF359944.1) from Lecythidaceae was used as the outgroup. Finally, IQtree v. 1.6 was used to construct a maximum likelihood tree with 1000× bootstrap resampling (Nguyen et al. Citation2015).
Results
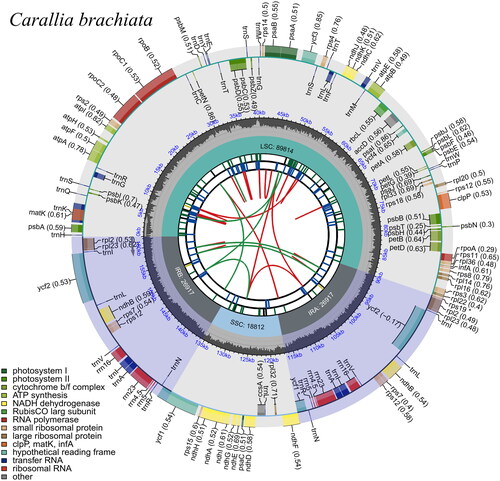
The complete chloroplast genome of C. brachiata was a typical quadripartite structure that contained 162,460 bp across four areas, including a large single copy (89,814 bp), a small single copy (18,804 bp) and a pair of inverted repeat regions (26,921 bp each) (). The total guanine + cytosine (GC) content of the genome was 35.76%. In total, 130 genes were annotated within the chloroplast genome of C. brachiata, including 85 PCGs, 37 tRNAs and 8 rRNA genes. Furthermore, 17 genes in the chloroplast genome of C. brachiata contained introns. Among them, trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA contained a single intron, whereas ycf3 and clpP had two introns. Owing to their location in the inverted repeat region, 12 genes were duplicated, including one PCG (ycf1), four rRNAs (rrn4.5, rrn5, rrn16 and rrn23), and seven tRNAs (trnI-CAU, trnL-CAA, trnA-UGC, trnI-GAU, trnV-GAC, trnR-ACG and trnN-GUU). Additionally, rps12 had three and two exons located on the inverted repeats, indicating that rps12 exhibited trans-splicing (supplemental Figure S1). Nine genes including atpF, rpoC1, clpP, petB, petD, rpl2, ndhB, ndhA, ndhB, and rpl2 are cis-splicing genes (supplemental Figure S2).
Figure 3. Circular representation of Carallia brachiata chloroplast genome, showing the clockwise (genes inside the circle) and counterclockwise (outside) transcribed genes. Colors identify genes from the same functional category, following the figure legends. In the inner circle, the dark and light grey bars indicate the guanine + cytosine and adenine + thymine content, respectively. IRa and IRb: inverted repeat regions; LSC: large single copy region; SSC: small single copy.
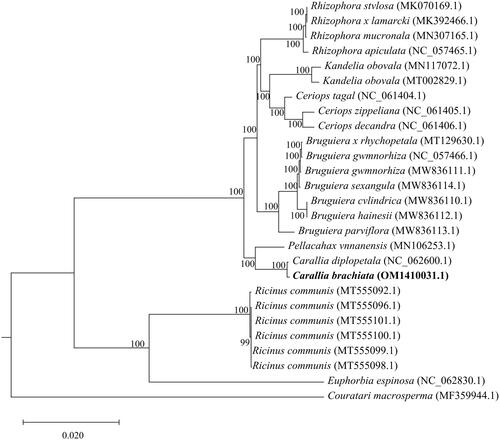
Twenty-nine species were initially used to construct the phylogenetic tree; however, the bootstrap value was too low to be valid; thus, the related species were removed. As a result, the final phylogenetic tree consisted of 27 species. The phylogenetic analysis revealed that C. brachiata was more closely related to Carallia diplopetala among all members of the Rhizophoraceae family ().
Figure 4. Maximum-likelihood phylogenetic tree for C. brachiata and 28 related species based on 62 homologous protein-coding genes. Bootstrap support values are indicated at each node (N = 1000). the scale bar indicates the phylogenetic distance in substitutions per site. The following sequences were used: Kandelia obovata (MN117072.1) (Du et al. Citation2019), Kandelia obovata (MT002829.1) (Xuli et al. 2020), Ceriops decandra (NC_061406.1) (Ruang-Areerate et al. Citation2022), Ceriops zippeliana (NC_061405.1) (Ruang-Areerate et al. Citation2022), Ceriops tagal (NC_061404.1) (Ruang-Areerate et al. Citation2022), Rhizophora stylosa (MK070169.1) (Li et al. Citation2019), Rhizophora × lamarcki (MK392466.1) (Zhang et al. Citation2019), Rhizophora mucronata (MN307165.1) (Wu Citation2019), Rhizophora apiculata (NC_057465.1) (Jiang Citation2020), Bruguiera gymnorhiza (NC_057466.1) (Jiang Citation2020), Bruguiera x rhynchopetala (MT129630.1) (Ying et al. Citation2020), Bruguiera gymnorhiza (MW836111.1) (Ruang-Areerate et al. Citation2021), Bruguiera sexangula (MW836114.1) (Ruang-Areerate et al. Citation2021), Bruguiera cylindrica (MW836110.1) (Ruang-Areerate et al. Citation2021), Bruguiera hainesii (MW836112.1)(Ruang-Areerate et al. Citation2021), Bruguiera parviflora (MW836113.1)(Ruang-Areerate et al. Citation2021), Carallia brachiata (OM141003.1) (this study), Carallia diplopetala (NC_062600.1) (Wang et al. Citation2021), Pellacalyx yunnanensis (MN106253.1) (Zhang et al. Citation2019), Ricinus communis (MT555096.1) (Muraguri et al. Citation2020), Ricinus communis (MT555101.1) (Muraguri et al. Citation2020), Ricinus communis (MT555100.1) (Muraguri et al. Citation2020), Ricinus communis (MT555099.1) (Muraguri et al. Citation2020), Ricinus communis (MT555098.1) (Muraguri et al. Citation2020), Ricinus communis (MT555092.1) (Muraguri et al. Citation2020), Euphorbia espinosa (NC_062830.1) (Wei Citation2021) and Couratari macrosperma (MF359944.1) (Vargas et al. Citation2017).
Discussion and conclusion
In this study, the complete chloroplast genome of C. brachiata was sequenced, revealing a total length of 162,460 bp, including four regions: a large single copy (89,814 bp), a small single copy (18,804 bp) and a pair of inverted repeats (26,921 bp each). The overall GC content was 35.76%. In total, 130 genes were annotated within the chloroplast genome, including 85 PCGs and 37 tRNA and 8 rRNA genes. Subsequent phylogenetic analyses revealed that C. brachiata is closely related to Carallia diplopetala (NC_062600.1). C. diplopetala, which exhibits the closest relationship to C. brachiata, has a slightly smaller chloroplast genome than C. brachiata, comprising 83 PCGs, 37 tRNAs and 8 rRNAs, with a total length of 162,052 bp (Wang et al. Citation2021).
Author contributions
You Zhou and Xiongmei Zhu performed the experiments, analyzed the data, authored drafts of the paper, and approved the final draft. Jiyun She analyzed the data, prepared the figures, and approved the final draft. Fen Xiao and Jian Zhao conceived and designed the experiment, reviewed the drafts of the paper, and approved the final draft. All authors agree to be accountable for all aspects of this study.
Ethical statement
Carallia brachiata leaves were collected from the South China Botanical Garden in Guangzhou, China in compliance with the national Wild Plant Protective Regulations.
Supplemental Material
Download TIFF Image (61.4 KB)Supplemental Material
Download TIFF Image (23.2 KB)Supplemental Material
Download PDF (316.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. OM141003. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA801906, SRR17823292, and SAMN25413005, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Du Z, Li J, Yang D. 2019. The complete chloroplast genome of a mangrove Kandelia obovata Sheue, Liu & Yong. Mitochondrial DNA B Resour. 4(2):3414–3415.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:10.1093/nar/gkh340.
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. doi:10.1186/s13059-015-0721-2.
- Jiang G-F. 2020. Bruguiera gymnorhiza chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Jiang G-F. 2020. Rhizophora apiculata chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Li C-T, Guo P, Huang H-R, Pei N-C, Shi M-M, Yan H-F. 2019. The complete chloroplast genome of Rhizophora stylosa and its phylogenetic implications. Mitochondrial DNA B Resour. 4(1):374–375. doi:10.1080/23802359.2018.1547167.
- Ling SK, Takashima T, Tanaka T, Fujioka T, Mihashi K, Kouno I. 2004. A new diglycosyl megastigmane from Carallia brachiata. Fitoterapia. 75(7–8):785–788. doi:10.1016/j.fitote.2004.09.019.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate E25 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate K314 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate K411 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate TB5 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate ZB306 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Muraguri S, Xu W, Chapman M, Muchugi A, Oluwaniyi A, Oyebanji O, Liu A. 2020. Ricinus communis isolate K32 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50. doi:10.1186/s13007-019-0435-7.
- Ruang-Areerate P, Kongkachana W, Naktang C, Sonthirod C, Narong N, Jomchai N, Maprasop P, Maknual C, Phormsin N, Shearman JR, et al. 2021. Complete chloroplast genome sequences of five Bruguiera species (Rhizophoraceae): comparative analysis and phylogenetic relationships. PeerJ. 9:e12268. doi:10.7717/peerj.12268.
- Ruang-Areerate P, Yoocha T, Kongkachana W, Phetchawang P, Maknual C, Meepol W, Jiumjamrassil D, Pootakham W, Tangphatsornruang S. 2022. Comparative analysis and phylogenetic relationships of ceriops species (Rhizophoraceae) and Avicennia lanata (Acanthaceae): insight into the chloroplast genome evolution between middle and seaward zones of mangrove forests. Biology (Basel). 11(3):383. doi:10.3390/biology11030383.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27(6):863–864. doi:10.1093/bioinformatics/btr026.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. doi:10.1080/10635150701472164.
- Vargas OM, Thomson AM, Dick CW. 2017. Couratari macrosperma voucher NY: Janovec2506 chloroplast, partial genome. Bethesda, MD: National Center for Biotechnology Information.
- Wang R, Liao N, Liu X, Qin Y, Xiao Y, Wang Y, Huang R. 2021. Carallia diplopetala chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Wei N. 2021. Euphorbia espinosa voucher CS 20151016-1 chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Wu H. 2019. Rhizophora mucronata plastid, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Xuli J, Tao L. 2020. Kandelia obovata chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Technology Information.
- Ying Z, Nian LX, Yong Y, Xiang J. 2020. Bruguiera × rhynchopetala chloroplast, complete genome. Bethesda, MD: National Center for Biotechnology Information.
- Zhang J, Li Y, Yuan X, Wang Y. 2019. The complete chloroplast genome sequence of Pellacalyx yunnanensis: an endangered species in China. Mitochondrial DNA B Resour. 4(2):3948–3949. doi:10.1080/23802359.2019.1688110.
- Zhang Y, Zhong J-D, Yuan C-C. 2019. Complete chloroplast genome of a Mangrove Natural Hybrid, Rhizophora × lamarckii. Mitochondrial DNA B Resour. 4(1):1465–1466. doi:10.1080/23802359.2019.1598790.