Abstract
The complete chloroplast genome sequence of Crepidomanes latealatum (Bosch) Copel. was determined in the present study. The genome is 145,943 base pairs (bp) in length and comprised two inverted repeats (32,990 bp) between a large single copy (92,170 bp) and a small single copy (20,783 bp). It contains 88 coding genes, 8 rRNA genes, 34 tRNA genes, and 1 pseudogene of trnL-UAA, and the GC content is 37.6%. Molecular phylogenetic analysis based on the plastid genome sequences of related taxa strongly supported the monophyly of the family Hymenophyllaceae, and the genus Vandenboschia was a sister group of Crepidomanes. In addition, compared to C. minutum, two large deletions of 453 bp and 878 bp were found in the IGS regions of petA-psbI and rrn16-trnV-GAC of C. latealatum cp genome, respectively.
Introduction
The genus Crepidomanes, which belongs to the family Hymenophyllaceae, comprises 30 species (PPG I, Citation2016). In the genus, Crepidomanes latealatum (Bosch) Copel. was described and published by Copeland (Citation1938) with a synonym Trichomanes latealatum Bosch and it has been treated as an accepted name in the present classification (Hassler et al. Citation2021). It is characterized by stipes very narrowly presence, ovate-oblong to oblong-subtriangular laminae, oblong and dark green pinnae, false veinlets presence, and cup-shaped and dilated involucres with campanulate in lower part and bilabiate in upper part (Lee et al. Citation2014), and is widely distributed from subtropical to tropical regions of Eurasia, such as Malaysia, India, Japan, China, and Korea (Moon Citation2008; Patil et al. Citation2014). However, in Korea, it is listed as a member of class V Floristic Target Species, which are distributed only in a very small area or are discontinuously distributed. Therefore, C. latealatum is regarded as an indicator of the conservation priority of major domestic species (Kim et al. Citation2018). Although complete chloroplast (cp) genome data for vascular plants have rapidly accumulated, a lack the information on ferns remains. In the present study, we clarified the cp genome structure of C. latealatum and compared it with related taxa to provide genomic data for understanding the phylogenetic relationships of the family Hymenophyllaceae.
Materials and methods
The plant material of C. latealatum was collected from Youngcheon-dong, Seogwipo-si, Jeju island, Korea (33°17’59.96"N, 126°34’47.31"E, alt. 288 m) (). A voucher specimen (CBNU2018-0153, [email protected]) was deposited at the Herbarium of Chungbuk National University (CBNU). Total genomic DNA was extracted from a leaf dehydrated using silica gel, using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Sequencing was conducted using the HiSeq X Ten system (Illumina, San Diego, CA, USA). In total, 17,554,992′ paired-end reads (151 base pairs (bp) in length) were obtained and trimmed using trimmomatic 0.39 (Bolger et al. Citation2014) with the options LEADING:10, TRAILING:10, SLIDINGWINDOW:4:20, and MINLEN:50. Then, 16,204,252 reads remained, which were assembled into the reference plastome of C. minutum (MN 905537), following Kim and Chase (Citation2017). Finally, a circular cp genome sequence was constructed with 330.2 coverage depth.
Figure 1. Photographs of Crepidomanes latealatum taken by S. H. Park at the sample collection site: (A) C. latealatum growing on a shady rock near a humid valley. (B) Winged rachis and tubular involucres with dilated lips of C. latealatum.

All genes were annotated based on existing genes in other fern cp genomes using Geneious 22.1.1 (Kearse et al. Citation2012). A cp genome map was generated using CPGView (Liu et al. Citation2023; http://www.1kmpg.cn/cpgview/). Each coding gene was aligned using MAFFT (Katoh et al. Citation2002). The concatenated alignment using 85 coding genes was 74,608 bp long. The partition models were selected using ModelFinder with edge-proportional partitions and merge options (Chernomor et al. Citation2016; Kalyaanamoorthy et al. Citation2017).
Maximum likelihood analysis was performed using IQ-Tree 2 (Minh et al. Citation2020) with 1,000 ultrafast bootstraps (Hoang et al. Citation2018). The IQTREE condition was iqtree2.exe -s Concatenated.phy -p partition_length.nex -m MFP + MERGE -bb 10000. Osmundastrum cinnamomeum (NC024157) was included in the phylogenetic analysis as an outgroup for related taxa of the family Hymenophyllaceae.
Results and discussion
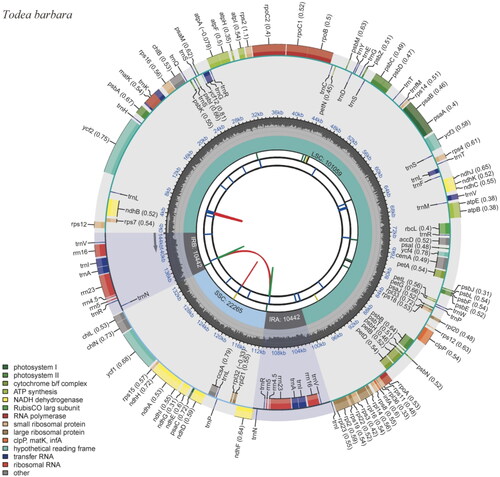
The cp genome of C. latealatum (NC072291, ) is 145,943 bp long with two inverted repeats (IRs) (32,990 bp) between a large single copy (LSC) (92,170 bp) and a small single copy (SSC) (20,783bp). It contains 131 genes, comprising 88 protein-coding genes, 8 ribosomal RNAs, 34 transfer RNAs, and 1 pseudogene of trnL-UAA. The GC content of the genome was 37.6% (Supplementary Figure 1). Compared to the cp genome of the related taxon C. minutum (MN 905537), it is 531 bp shorter in length and 0.5% higher in GC content. In addition, two large deletions of 453 bp and 878 bp occurred in the IGS regions of petA-psbI and rrn16-trnV-GAC, respectively. However, another large deletion of 3,100 bp was observed in the maK-rps16 region of the C. minutum cp genome.
Figure 2. Complete chloroplast genome map of Crepidomanes latealatum, containing six tracks. From the center, the first track shows the dispersed repeats, which consist of direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, LSC, SSC, and IRs, are identified on the fourth track. The GC content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
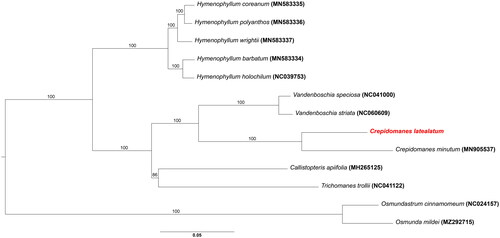
The molecular phylogenetic analysis strongly supported the monophyly of the family Hymenophyllaceae, and a sister relationship between Crepidomanes and Vandenboshia was also confirmed within the family (). It was consistent with previous phylogenetic study of Ebihara et al. (Citation2006).
Figure 3. Phylogenetic tree of Crepidomanes latealatum and related taxa inferred from maximum likelihood analysis based on the 88 coding genes sequences. The outgroup was Osmundastrum chinnamomeum (NC 024157; Kim et al. Citation2014) and 5 genera of the family Hymenophyllaceae were included together with osmunda mildei (MZ292715). the bootstrap supporting values are described on the branches. The position of C. latealatum is indicated in red. The following sequences were used: MN583336 for hymenophyllum polyanthos (Kim and Kim Citation2020), MN583335 for H. coreanum (Kim and Kim Citation2020), MN583337 for H. wrightii (Kim and Kim Citation2020), MN039753 for H. holochilum (Kuo et al. Citation2018), NC063573 for Crepidomanes minutum, NC041000 for Vandenboschia speciosa (Ruiz-Ruano et al. Citation2019), NC060609 for V. striata (Wang et al. Citation2022), MH265125 for callistopteris apiifolia, and NC041122 for Trichomanes trollii (Lehtonen Citation2018).
Conclusion
The complete cp genome of Crepidomanes latealatum is presented for the first time in this study. It has a typical circular form composed of 145,943 bp and 132 genes. Based on molecular phylogenetic analysis using the cp genome sequence data, the phylogenetic position of Crepidomanes latealatum was confirmed in the family Hymenophyllaceae, and the genus Crepidomanes was shown to be a sister group of Vandenboschia.
Authors’ contributions
Conceiving and designing the study, JSK and HTK; collecting the plant material, SHP and HTK; managing the voucher specimen and extracting genomic DNA, SHP; performing and analyzing data, JHH and HTK; writing – original draft preparation, JHH; writing – review and editing, JSK; supervision, JSK and HTK; all authors have read and agreed to the published version of the manuscript.
Ethical approval
Crepidomanes latealatum is not an endangered or protected species; therefore, permission is not required to collect this species. Research on this species, including the collection of plant materials, was conducted following the guidelines provided by the Chungpook National University.
Supplemental Material
Download JPEG Image (2.3 MB)Supplemental Material
Download JPEG Image (7 MB)Data availability statement
The genome sequence data supporting the findings of this study are available in the NCBI GenBank under accession no. NC072291 (https://www.ncbi.nlm.nih.gov/nuccore/ NC_072291.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA951456, SRR24044269, and SAMN34043022, respectively.
Disclosure statement
The authors have no conflicts of interest in this study.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformat. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. doi: 10.1093/sysbio/syw037.
- Copeland T. 1938. Genera Hymenophyllacearum. Philipp J Sci. 67(1):60–61.
- Ebihara A, Dubuisson J-Y, Iwatsuki K, Hennequin S, Ito M. 2006. A taxonomic revision of Hymenophyllaceae. Blum J Plant Tax Plant Geog. 51(2):221–280. doi: 10.3767/000651906X622210.
- Hassler M. 2021. Checklist of Ferns and Lycophytes of the World. In O. Bánki, Y. Roskov, L. Vandepitte, R. E. DeWalt, D. Remsen, P. Schalk, T. Orrell, M. Keping, J. Miller, R. Aalbu, R. Adlard, E. Adriaenssens, C. Aedo, E. Aescht, N. Akkari, M. A. Alonso-Zarazaga, B. Alvarez, F. Alvarez, G. Anderson, et al., Catalogue of Life Checklist (Version 2021-08-06).
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformat. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kim CH, Moon MO, Ahn JG, Hwang IC, Lee SH, Choi SS, Bum HM, Cha JY. 2018. Floristic target species (FT species) in Korea. National Institute of Ecology Press, Seocheon, Korea; p. 728. (in Korean)
- Kim HT, Chase MW. 2017. Independent degradation in genes of the plastid NDH gene family in species of the orchid genus Cymbidium (Orchidaceae: Epidendroideae). PLoS One. 12(11):e0187318. doi: 10.1371/journal.pone.0187318.
- Kim HT, Chung MG, Kim KJ. 2014. Chloroplast genome evolution in early diverged leptosporangiate ferns. Mol Cells. 37(5):372–382. doi: 10.14348/molcells.2014.2296.
- Kim HT, Kim JS. 2020. The dynamic evolution of mobile open reading frames in plastomes of Hymenophyllum Sm. and new insight on Hymenophyllum coreanum Nakai. Sci Rep. 10(1):11059. doi: 10.1038/s41598-020-68000-7.
- Kuo LY, Qi X, Ma H, Li FW. 2018. Order-level fern plastome phylogenomics: new insights from Hymenophyllales. Am J Bot. 105(9):1545–1555. doi: 10.1002/ajb2.1152.
- Lee CS, Lee K, Hwang Y. 2014. First record of Crepidomanes schmidtianum (Zenker ex Tasch.) K. Iwats. (Hymenophyllaceae) from Korea. Korean J Plant Taxonomy. 44(1):1–5. doi: 10.11110/kjpt.2014.44.1.1.
- Lehtonen S. 2018. The complete plastid genome sequence of Trichomanes trollii (Hymenophyllaceae). Nordic J. Bot. 36:e02072.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):1–11.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Moon MO. 2008. Pteridophyte Flora of Jeju Island, Korea [Doctoral dissertation]. Jeju National University.
- Patil S, Hande P, Dongare M. 2014. A Filmy Fern: Crepidomanes latealatum (bosch) Copel. New to Northern Western Ghats of India. Indian Forester. 140(9):939–940.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54(6):563–603.
- Ruiz-Ruano FJ, Navarro-Domínguez B, Camacho JPM, Garrido-Ramos MA. 2019. Full plastome sequence of the fern Vandenboschia speciosa (Hymenophyllales): structural singularities and evolutionary insights. J Plant Res. 132(1):3–17. doi: 10.1007/s10265-018-1077-y.
- Wang R, Zhao G, Wang Z, Lin Z, Jiang X, Lin S, Chan H, Hong Y, Zhang Y, Su Y, et al. 2022. The complete chloroplast genome of Vandenboschia striata, a common and widespread filmy fern (Hymenophyllaceae. Mitochondrial DNA B Resour. 7(1):128–129. doi: 10.1080/23802359.2021.2013744.
