Abstract
In this study, we determined the chloroplast genome sequence of the Austral king fern, Todea barbara (L.) Moore. The plastome of T. barbara is a typical circular form composed of 144,208 bp with two inverted repeats (IRs; 10,442 bp), a large single copy (LSC; 101,059 bp), and a small single copy (SSC; 22,265 bp). The complete sequence comprises 131 genes, namely 85 protein-coding genes, eight ribosomal RNAs, and 38 transfer RNAs. The guanine–cytosine (GC) content of the genome was found to be 39.9%. Additionally, U-to-C RNA editing sites were identified in eight genes: atpE, chlB, clpP, matK, rpl20, rpoB, rpoC1, and rpoC2. Phylogenetic analysis using 85 coding gene sequences revealed that the genera Todea and Osmunda form a clade and that the genus Osmundastrum is a sister genus to both.
Introduction
Todea Willd. ex Bernh. is a genus of the family Osmundaceae and consists of only two species: Todea barbara (L.) Moore 1857 and T. papuana Hennipman 1968 (PPG I Citation2016). Unlike T. papuana, which occurs only in Papua New Guinea, T. barbara, known as the Austral king fern, is widely distributed; it is found in New Zealand, eastern and southeastern Australia, and southern Africa (Kato Citation2007).
T. barbara is an arborescent fern with a single trunk that grows to be massive with vegetative buds and develops a large root mantle. The fronds are large and coriaceous. Sporangia are restricted to the lower pinnules of the primary pinnae, especially in the lower part of the frond, and they are massed along the minor veinlets (; Flora of Australia Citation2023, http://www.ausflora.org.au).
Figure 1. Photographs of Todea barbara captured by Ki-Joong Kim at the collection site. The Austral king fern has a barrel-shaped trunk covered in dark brown to black fibrous aerial roots. The stipes are smooth and yellow-brown, with ear-like lobes at the base. The fronds are large and coriaceous.

The first chloroplast (cp) genome reported from the family Osmundaceae, obtained from Osmundatrum cinnamomeum (L.) C.Presl, showed that the cp genome feature of this early diverging leptosporangiate fern family was similar to that of eusporangiate ferns (Kim et al. Citation2014). However, only two complete chloroplast (cp) genomes of Osmunda species have been sequenced, and there is, to date, no information on the remaining four genera within Osmundaceae (Claytosmunda, Leptopteris, Plenasium, and Todea). Accordingly, we sequenced the complete cp genome of T. barbara and analyzed the phylogenetic relationships of this species within the family Osmundaceae.
Materials and methods
A specimen of Todea barbara was collected from the Royal Botanical Garden Sydney, Sydney, Australia (33° 86′ 41.24″ S, 151° 21′ 67.96″ E) in January 2019 by Prof. Ki-Joong Kim of the Korea University and was dispensed at the Plant DNA Bank in Korea (PDBK, https://pdbk.korea.ac.kr/), located in Seoul, according to the dispensing procedure of the PDBK. In addition, the specimen was deposited at the Korea University Herbarium (https://pdbk.korea.ac.kr/, contact to Ki-Joong Kim, [email protected]) under the voucher number TA2019-0031.
High-throughput sequencing of a sample from the collected specimen was performed using an Illumina Hiseq X-ten sequencing platform. In total, 17,563,188 paired-end raw reads (151 bp in length) were generated, and the raw data were trimmed using Trimmomatic 0.39 (Bolger et al. Citation2014), with the options LEADING:10, TRAILING:10, SLIDINGWINDOWS:4:20, and MINLEN:50. The complete cp genome was assembled into the reference genome of Osmunda japonica (MK554796) using 17,539,752 trimmed reads and annotated using the protocol described by Kim and Chase (Citation2017), except for the read-trimming stage. Lastly, the cp genome sequence was constructed with 477.2 coverage depth and submitted to the NCBI database under the accession number OP793887. In addition, a cp genome map was generated using CPGView (Liu et al. Citation2023; http://www.1kmpg.cn/cpgview/).
To perform a phylogenetic analysis of the family Osmundaceae, the cp genomes of three species (Osmunda japonica, MK554796; Osmunda mildei, MZ292715; and Osmundastrum cinnamomeum, NC024157) were utilized, and three cp genomes from Angiopteris (downloaded from GenBank) were treated as outgroups. Each coding gene was aligned using MAFFT (Katoh et al. Citation2002). The concatenated alignment using 85 coding genes was 74,123 bp long. Partition models were chosen using ModelFinder with edge-proportional partitions and merge options (Chernomor et al. Citation2016; Kalyaanamoorthy et al. Citation2017). A phylogenetic tree based on maximum likelihood analysis was constructed using IQ-TREE 2 (Minh et al. Citation2020) with 10,000 ultrafast bootstraps (Hoang et al. Citation2018). The IQ-TREE condition was iqtree2.exe-s Concatenated.phy-p partition_length.nex-m MFP + MERGE-bb 10000.
Results and discussion
The complete cp genome of Todea barbara (OP793887, ) is 144,208 bp in length, with two inverted repeats (IRs; 10,442 bp) between a large single copy (LSC; 101,059 bp) and a small single copy (SSC; 22,265 bp). It is longer than other Osmundaceae cp genomes recorded to date () and comprises 131 genes, namely 85 protein-coding genes, eight ribosomal RNAs, and 38 transfer RNAs. The guanine–cytosine (GC) content was 39.9%. Contrary to the other documented cp genomes of the family Osmundaceae (Osmunda and Osmundastrum), which have been shown to be similar to those of eusporangiate ferns in terms of restricted RNA editing sites (Wolf et al. Citation2004; Kim et al. Citation2014; Guo et al. Citation2015), eight genes in the cp genome of T. barbara, namely atpE, chlB, clpP, matK, rpl20, rpoB, rpoC1, and rpoC2, are likely to have internal stop codons () even it was not experimentally proved. Because these genes are generally conserved in the cp genomes of monilophytes, U-to-C RNA editing events are thought to occur independently in T. barbara within the family. Although C-to-U RNA editing occurs more often in land plants, U-to-C RNA editing has been observed in hornwort and monilophyte organelles (Knie et al. Citation2016).
Figure 2. Complete chloroplast (cp) genome map of Todea barbara, containing six tracks. From the center, the first track shows the dispersed repeats, which consist of direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, LSC, SSC, and IRs, are identified on the fourth track. The GC content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
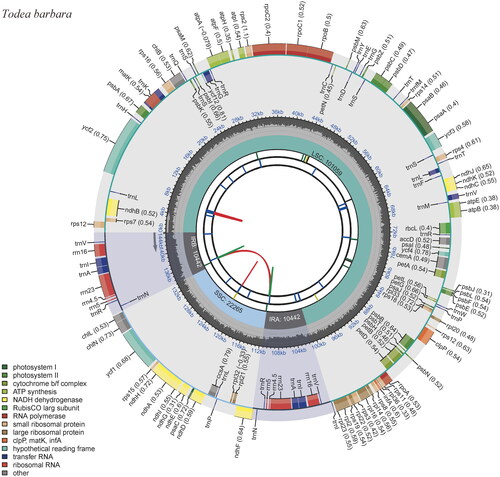
Figure 3. The internal stop codons in eight genes of Todea barbara. The numbers above the bases indicate the positions of the gene.

Table 1. Comparison of the complete chloroplast genomes of Todea and its related species used in the present study.
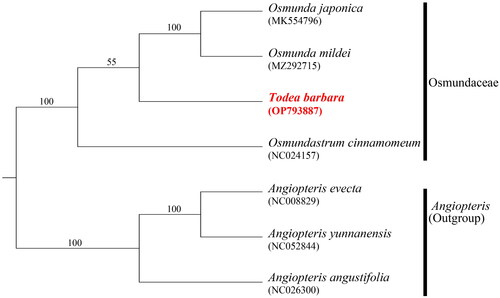
A phylogenetic tree of the family Osmundaceae, constructed using 85 coding gene sequences, revealed that the genera Todea and Osmunda form a clade in the family, and the genus Osmundastrum is sister to them (). This finding is consistent with the previously reported phylogenetic relationships among genera in the family Osmundaceae, as determined by analyzing seven cp genes (Metzgar et al. Citation2008). However, the low bootstrap value for the clade composed of Todea and Osmunda appears to be due to the restricted number of taxa, in particular, the lack of taxa closely related to Leptoptaris, which is regarded as a sister to Todea (Yatabe et al. Citation1999; Jud et al. Citation2008; Carvalho et al. Citation2013).
Figure 4. Maximum likelihood tree based on the 85 cp coding genes. Numbers on the nodes represent the bootstrap support values. The outgroups were three Angiopteris of A. evecta (NC024157; Roper et al. Citation2007), A. yunnanensis (NC052844; Jiang et al. Citation2019), and A. angustifolia (NC026300). Two Osumnda species (O. japonica; MK554796, Xu et al. Citation2019 and O. mildei; MZ292715) and Osmundatrum cinnamomeum (NC024157; Kim et al. Citation2014) were also included for the molecular phylogenetic analysis of the family Osmundaceae.
Conclusions
The complete cp genome of Todea Barbara, investigated for the first time in this study, was found to have a typical circular form composed of 144,208 bp and 131 genes. Based on molecular phylogenetic analysis using 85 coding gene sequences of cp genome, we determined that the genera Todea and Osmunda form a clade in the family, and the genus Osmundastrum is sister to them. In addition, U-to-C RNA editing sites were observed in eight genes: atpE, chlB, clpP, matK, rpl20, rpoB, rpoC1, and rpoC2 in the cp genome of T. barbara.
Ethical approval
Todea barbara is not included in the list of Australian threatened plants (https://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl?wanted=flora) and was collected with permission from the Royal Botanical Garden Sydney. Research on this species, including the collection of plant materials, was conducted following the guidelines provided by Korea University and Kyoungpook National University.
Author contributions
Conceiving and designing the study, JSK and HTK; performing and analyzing data, HTK; writing the original draft, JSK and HTK; review and editing, JSK; supervision, JSK. All authors have read and agreed to the published version of the manuscript.
Acknowledgment
The authors express their gratitude to Prof. Ki-Joong Kim of Korea University, Seoul, Korea, for providing the plant material essential for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting the findings of this study are available from NCBI GenBank under accession no. OP793887 (https://www.ncbi.nlm.nih.gov/nuccore/OP793887.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA954512, SRR24137031, and SAMN34146952, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Carvalho MR, Wilf P, Hermsen EJ, Gandolfo MA, Cúneo NR, Johnson KR. 2013. First record of Todea (Osmundaceae) in South America, from the early Eocene paleorainforests of Laguna del Hunco (Patagonia, Argentina). Am J Bot. 100(9):1831–1848. doi: 10.3732/ajb.1200637.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. doi: 10.1093/sysbio/syw037.
- Flora of Australia. 2023. Australian biological resources study, Canberra. [accessed 2023 Apr 10]. http://www.ausflora.org.au.
- Guo W, Grewe F, Mower JP. 2015. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. PLoS One. 10(1):e0117075. doi: 10.1371/journal.pone.0117075.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281.
- Jiang Q, Schneider H, Liu H. 2019. Complete chloroplast genome of Angiopteris yunnanensis (Marattiaceae). Mitochondrial DNA B Resour. 4(2):3912–3913. doi: 10.1080/23802359.2019.1688111.
- Jud NA, Rothwell GW, Stockey RA. 2008. Todea from the Lower Cretaceous of western North America: implications for the phylogeny, systematics, and evolution of modern Osmundaceae. Am J Bot. 95(3):330–339. doi: 10.3732/ajb.95.3.330.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Kato M. 2007. Distribution of Osmundaceae. Bull Natl Mus Nat Sci Ser B Bot. 33:81–90.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Kim HT, Chase MW. 2017. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS One. 12(11):e0187318. doi: 10.1371/journal.pone.0187318.
- Kim HT, Chung MG, Kim KJ. 2014. Chloroplast genome evolution in early diverged leptosporangiate ferns. Mol Cells. 37(5):372–382. doi: 10.14348/molcells.2014.2296.
- Knie N, Grewe F, Fischer S, Knoop V. 2016. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns – a monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol Biol. 16(1):134. doi: 10.1186/s12862-016-0707-z.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Eco Res. 23(3):694–704.
- Metzgar JS, Skog JE, Zimmer EA, Pryer KM. 2008. The paraphyly of Osmunda is confirmed by phylogenetic analyses of seven plastid loci. Syst Bot. 33(1):31–36. doi: 10.1600/036364408783887528.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54(6):563–603.
- Roper JM, Hansen SK, Wolf PG, Karol KG, Mandoli DF, Everett KDE, Kuehl J, Boore JL. 2007. The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). Am Fern J. 97(2):95–106. doi: 10.1640/0002-8444(2007)97[95:TCPGSO.2.0.CO;2]
- Wolf PG, Rowe CA, Hasebe M. 2004. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 339:89–97. doi: 10.1016/j.gene.2004.06.018.
- Xu L, Xing Y, Wang B, Liu C, Wang W, Kang T. 2019. Plastid genome and composition analysis of two medical ferns: Dryopteris crassirhizoma Nakai and Osmunda japonica Thunb. Chin Med. 14:9. doi: 10.1186/s13020-019-0230-4.
- Yatabe Y, Nishida H, Murakami N. 1999. Phylogeny of Osmundaceae inferred from rbcL nucleotide sequences and comparison to the fossil evidences. J Plant Res. 112(4):397–404. doi: 10.1007/PL00013894.
