Abstract
Amorphophallus coaetaneus S. Y. Liu & S. J. Wei 1986 is a perennial herb belonging to the Araceae family in southwestern China (Guangxi and Yunnan provinces). Although this species have not been list in the red list of International Union for Conservation of Nature (IUCN), the populations are declining due to human over exploitation. To help to genetic diversity studies, we sequenced and assembled the complete chloroplast (cp) genome of A. coaetaneus (GenBank accession number of national center for biotechnology information (NCBI): OQ404947). The assembled genome revealed 175,465 bp in length with a GC content of 34.90%, including a large single-copy (LSC) region (98,561 bp), a small single-copy (SSC) region (16,504 bp) and two inverted repeat regions (IRs) (30,200 bp each). A total of 133 genes were annotated, of which 85 are protein-coding genes, 40 are tRNA genes and 8 are rRNA genes. As an output of this study, a maximum likelihood (ML) phylogenetic inference of 16 Araceae species clustered all four Amorphophallus species in one clade, and showed a relatively close relationship between the tribes Pythonieae and Colocasieae. The cp genome will serve as a basis in a more extensive molecular works covering all possible extant population of Amorphophallus, as well as conservation, breeding, and other ethnobotanical utilization of this species.
Introduction
Among all terrestrial plants, Araceae is one of the most diversified groups of the Angiosperm families with a high species richness and phenotypic variations. This family includes the smallest angiosperm (Wolffia Horkel ex Schleid) as well as the species with the largest flower in the world (Amorphophallus titanum) (Pouchon et al. Citation2023). Amorphophallus coaetaneus S. Y. Liu & S. J. Wei 1986 is a perennial herb that belongs to the Amorphophallus genus of the Araceae family, and was first discovered in Guangxi province, China in 1986 (Li et al. Citation2010). The natural habitat of this species is moist forested valleys, watersides, and thickets in a geographic region including Guangxi and Yunnan provinces, China (Li et al. Citation2010). It is notably known that the plant species can accumulate a large amount of konjac glucomannan (KGM), and several Amorphophallus species have a long history of cultivation and for use as food and medicine in many Asian countries, such as China, Japan, and India (Srzednicki and Borompichaichartkul Citation2020). Although most of Amorphophallus species in China have not been list in the red list of International Union for Conservation of Nature (IUCN), the wild resources are declining due primarily to the loss of its natural habitat (Yin et al. Citation2021). Genetic diversity and phylogeographic studies are needed for the conservation of natural populations. Due to its characteristics of monophyletic inheritance and having no recombination, the chloroplast genome has been considered an ideal tool for phylogeographic studies (Mehmood, Abdullah, Shahzadi et al. Citation2020, Mehmood, Abdullah, Ubaid et al. Citation2020). In this study, we sequenced and assembled the complete chloroplast (cp) genome of A. coaetaneus, which will aid in the conservation and further utilization of members of the genus Amorphophallus.
Materials
The leaf sample of one individual of A. coaetaneus was collected from Xishuangbanna, Yunnan province, China (Longitude: 101°29’46.03" E; Latitude: 21°31’54.95" N) (). Genomic DNA was isolated using a commercial plant DNA isolation kit (DP305; Tiangen, Beijing), and was then stored in a −80 °C freezer in the molecular laboratory of Qujing Normal University. The specimen (MLMH-1-20220722QJNU; Yong Gao, 22 July, 2022) was deposited in the herbarium of Qujing Normal University (Yong Gao, [email protected]).
Methods
Whole genome sequencing of A. coaetaneus was conducted using the Illumina NovaSeq platform (Illumina, California) by Novogene (Beijing). The genomic DNA was fragmented randomly, and fragments of approximately 350 bp were selected for paired-ended sequencing with a length of 150 bp. The raw sequencing data were first filtered and then assembled with the software Getorganelle v1.7.6 (Jin et al. Citation2020). We used default parameters except that k-mers were set to75, 95, 115, and 127. The genomic features of the assembled chloroplast genome were annotated with the online software CPGAVAS2 (http://47.96.249.172:16019/analyzer/home) (Shi et al. Citation2019) and GeSeq using default parameters (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017). In addition, the OGDRAW software was utilized to generate a circular genome map (Greiner et al. Citation2019). To infer the phylogenetic positon of A. coaetaneus, cp genomes of 15 Araceae species were downloaded from GenBank of national center for biotechnology information (NCBI) (Bethesda, MD, USA), and the DNA sequences of the protein-coding genes were extracted. The gene sequences were aligned using MAFFT v7.475 and then concatenated into one sequence (Katoh and Standley Citation2013). Spirodela polyrhiza (L.) Schleid was chosen as an outgroup for the phylogenetic analysis. The optimal nucleotide substitution model was identified by ModelFinder (Kalyaanamoorthy et al. Citation2017), and a maximum likelihood (ML) phylogenetic tree was constructed using iqtree v1.6.12 with 1000 bootstraps (Nguyen et al. Citation2015).
Results
As a result of the study, the complete genome assembly of A. coaetaneus was revealed for the first time. A total of 9.02 Gb of raw data was produced, and the assembled cp genome was found to be 175,465 bp in length with a typical quadripartite structure. The overall GC content of the genome was 34.90%. Lengths of the large single-copy (LSC) region, small single-copy (SSC) region, and two inverted repeat regions (IRs) were 98,561 bp, 16,504 bp, 30,200 bp, and 30,200 bp, respectively (). In total, 133 genes were annotated, including 85 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. Among these genes, 16 genes (tRNA-Ala, tRNA-Gly, tRNA-Ile, tRNA-Leu, tRNA-Lys, tRNA-Val, rpl16, rpl2, rps16, rpoC1, accD, atpF, petB, petD, ndhA and ndhB) had one intron, and two genes (ycf3 and clpP) had two introns. The TVM + F+R2 model was chosen as the best-fit nucleotide substitution model according to Bayesian Information Criterion. The phylogenetic tree clustered all four Amorphophallus species into one clade with a support value of 100% (). This tree suggested a close relationship between A. albus and A. konjac K. Koch. In addition, the genera Alocasia, Caladium, and Syngonium were shown to be closely related to Amorphophallus. The assembled chloroplast genome sequence of A. coaetaneus was deposited to NCBI GenBank with accession number OQ404947.
Figure 2. The circular map of the chloroplast genome from A. coaetaneus (OQ404947). genes belonging to diferent functional groups are plotted in the outer circle. The quadripartite structure including LSC, SSC, IRA and IRB is shown. The dark gray in the inner circle indicates GC content of the genome.
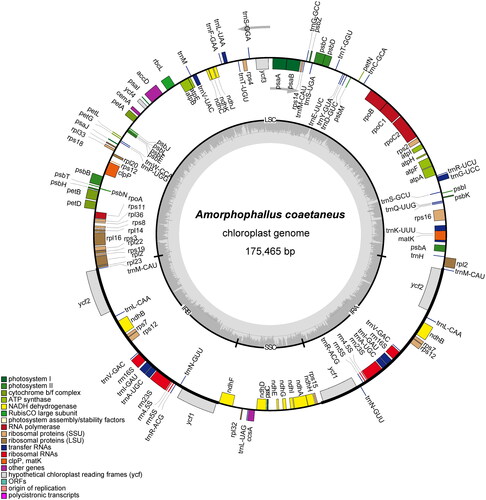
Figure 3. Maximum-likelihood phylogenetic tree of 15 related taxa and the placement of A. coaetaneus. Spirodela polyrhiza is used as an outgroup. The bootstrap support values are shown at each node. Accession numbers: Amorphophallus albus, NC_067990 (reference not available); Amorphophallus konjac, MK611803 (Hu et al. Citation2019); Amorphophallus coaetaneus, OQ404947 (this study); Amorphophallus titanium, NC_056329 (reference not available); Alocasia cucullata, NC_060696 (Low et al. Citation2021); Caladium bicolor, NC_060474 (reference not available); Syngonium angustatum, MN046894 (Henriquez et al. Citation2020a); Typhonium giganteum, MN626718 (Kim et al. Citation2020); Typhonium trifoliatum, MW451769 (reference not available); arisaema decipiens, NC_064687 (reference not available); pinellia cordata, MT863558 (reference not available); carlephyton glaucophyllum, MT161478 (reference not available); montrichardia arborescens, MN046889 (Henriquez et al. Citation2020a); epipremnum amplissimum, NC_047232 (Henriquez et al. Citation2020b); monstera adansonii, MN046888 (Henriquez et al. Citation2020b); Spirodela polyrhiza, MN419335 (reference not available).

Discussion and conclusion
Our phylogenetic study of 16 Araceae species suggests a relatively close relationship between the tribes Pythonieae and Colocasieae within the subfamily Aroideae, which improves our understanding of the phylogenetic relationships within Araceae. Previous phylogenetic studies of the Amorphophallus genus based on nuclear (ITS1) and cpDNA (rbcL and matK) sequences found four subgenera (Scutandrium, Amorphophallus, Metandrium and Afrophallus) within 157 species (Claudel et al. Citation2017). A. albus and A. konjac, which were assigned in the same subgenus (Scutandrium) in the previous study, were found to be sister groups with a bootstrap value of 100% in our phylogenetic analysis. Besides, our phylogenetic tree with additional species also revealed a close relationship between A. coaetaneus and A. titanium.
Amorphophallus is a genus with more than 200 species (Mayo et al. Citation1997). However, only a handful of complete cp genomes have been reported to date, which has hindered studies of population genetics (Hu et al. Citation2019). In this study, we sequenced, assembled and annotated the cp genome of A. coaetaneus. In the context of the high economic value of KGM, this cp genomic resource from A. coaetaneus will aid in the development of molecular markers in the future and will further aid in the species delineation, molecular breeding of high KGM varieties, and genetic diversity studies.
Ethical approval
This study includes no human, animal, or endangered plant samples, and the sample was collected legally following guidelines provided by the authors’ institution and national or international regulations.
Author contributions
Yong Gao performed the molecular experiment. Yin Si and Kun Dong analyzed the data. Penghui Xiao and Weijia Wu collected the sample. Yong Gao wrote the manuscript. All authors have revised and approved the final version of this manuscript.
Supplemental Material
Download MS Word (146.1 KB)Acknowledgments
The authors thank Dr. Joseph Elliot at the University of Kansas for her assistance with English language and grammatical editing of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome of Amorphophallus coaetaneus was deposited to the national center for biotechnology information (NCBI) (Bethesda, MD 20894, USA) GenBank database (https://www.ncbi.nlm.nih.gov/) under the accession number OQ404947. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA943846, SAMN33734031, and SRR23825299, respectively.
Additional information
Funding
References
- Claudel C, Buerki S, Chatrou LW, Antonelli A, Alvarez N, Hetterscheid W. 2017. Large-scale phylogenetic analysis of Amorphophallus (Araceae) derived from nuclear and plastid sequences reveals new subgeneric delineation. Botan J Linnean Soc. 184(1):32–45. doi:10.1093/botlinnean/box013.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Henriquez CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. 2020a. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 112(3): 2349–2360. doi:10.1016/j.ygeno.2020.01.006.
- Henriquez, CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. 2020b. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae). Planta. 251(3): 72. doi:10.1007/s00425-020-03365-7.
- Hu H, Liu J, Wang B, An J, Wang Q. 2019. Characterization of the complete chloroplast genome of Amorphophallus konjac (Araceae) and its phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1658–1659. doi:10.1080/23802359.2019.1606683.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong T, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kim J, Lee J, Choi S, Um S, Kim H, Chun HS, Nah G. 2020. The complete chloroplast genome of Typhonium giganteum (Araceae). Mitochondrial DNA B Resour. 5(3):2994–2995. doi:10.1080/23802359.2020.1797571.
- Li H, Zhu G, Boyce PC, Jin M, Hetterscheid WLA, Bogner J, Jacobsen N. 2010. Araceae. Flora of China. Beijing: science Press.
- Low SL, Yu C-C, Ooi IH, Eiadthong W, Galloway A, Zhou Z-K, Xing Y-W. 2021. Extensive Miocene speciation in and out of Indochina: the biogeographic history of Typhonium sensu stricto (Araceae) and its implication for the assembly of Indochina flora. J Syst Evol. 59(3):419–428. doi:10.1111/jse.12689.
- Mayo SJ, Bogner J, Boyce PC. 1997. The genera of Araceae. Kew: Royal Botanic Gardens.
- Mehmood F, Abdullah, Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2): 1522–1530. doi:10.1016/j.ygeno.2019.08.024.
- Mehmood F, Abdullah, Ubaid Z, Bao Y, Poczai P, Mirza B. 2020. Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9(6): 752. doi:10.3390/plants9060752.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 32(1):268–274. doi:10.1093/molbev/msu300.
- Pouchon C, Gauthier J, Pitteloud C, Claudel C, Alvarez N. 2023. Phylogenomic study of Amorphophallus (Alismatales; Araceae): When plastid DNA gene sequences help to resolve the backbone subgeneric delineation. J of Sytematics Evolution. 61(1):64–79. doi:10.1111/jse.12910.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Srzednicki G, Borompichaichartkul C. 2020. Konjac Glucomannan-Production, Processing, and Functional Applications. Boca Raton: CRC Press.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Yin S, Chu H, Zhang Y, Lu F, Gao Y. 2021. Population genetic diversity and genetic structure of Amorphophallus yunnanensis in Southwestern China and its conservation implication. Taiwania. 66(2):126–134.