Abstract
Grewia biloba var. parviflora (Bunge) Hand.-Mazz. (1933), a shrub or small tree, is native to northern and southern China. It is an excellent relief and medicinal plant. The complete chloroplast genome is 158,043 bp in length, with a large single-copy region of 86,957 bp, a small single-copy region of 20,138 bp, two inverted repeat regions of 25,474 bp each, and a GC content of 37.4%. There were 129 genes annotated, including 84 known protein-coding genes, 37 tRNAs, and eight rRNAs. The phylogenetic trees are constructed using plastome data from 38 species and the maximum-likelihood method. The results of the chloroplast genome-wide analysis and the phylogenetic tree show the taxonomic phylogeny of the G. biloba var. parviflora in relation to other species, increasing the accuracy of the phylogenetic classification of the plant.
Introduction
Grewia biloba var. parviflora (Bunge) Hand.-Mazz. (1933), a shrub or small tree, is native to northern and southern China, according to The Flora of China (Li and Hedge Citation1933). There are several medicinal aliases for G. biloba var. parviflora in China, such as ‘kid’s fist’, ‘Ge Fei Ma’, ‘cotton tendon strip’, etc. (Xie Citation2004). The mature G. biloba var. parviflora has a well-developed stem and bark, and the mature fruits are non-toxic and edible. In ancient China, it was often used as a folk remedy for tonics, deficiencies, and kidneys. At this stage, most of the research on G. biloba var. parviflora is focused on its chemical composition and the pharmacological function of the extracts as anti-tumor and anti-malarial (Liu et al. Citation2009). In Sudan, in north-eastern Africa, it is used as a sedative to detach human and bovine placenta (Chen Citation1989). In addition, as a drought-tolerant plant, G. biloba var. parviflora’s leaves and secondary xylem exhibit a high degree of plasticity in their anatomical and physiological characteristics (Shi et al. Citation2006), so scholars are mostly interested in their water use characteristics and community characteristics under drought stress (Sun et al. Citation2014; Zhang Citation2021).
Materials and methods
Fresh leaves were collected from Jinzhou City, Liaoning Province, China (E121°02′34.41″, N41°10′16.55″) (). The specimen voucher was identified by Professor Tingguo Kang from the Liaoning University of Traditional Chinese Medicine in Shenyang, China. A specimen was deposited at the herbarium of Liaoning University of Traditional Chinese Medicine (Liang Xu [email protected], G. biloba var. parviflora number: 210711191005014LY) (Supplemental Figure S4). All operations were carried out according to the Specification on Good Agriculture and Collection Practices for Medicinal Plants (GACP; number: T/CCCMHPIE 2.1-2018).
Figure 1. Photographs of (A) a flowering branch and (B) a leaf blade of G. biloba var. parviflora taken by Zhang Shumei in Dalian, Liaoning, China (E121°35′49″, N38°58′11″). The leaves are densely covered with soft yellow-brown hairs on the underside, and the flowers are short and small.

Total genomic DNA was extracted from 150 mg fresh leaves using the cetyltrimethylammonium bromide method (Doyle and Doyle Citation1987). An aliquot of purified DNA (1 μg) was fragmented by sonication to a size of 350 bp, then to construct a short-insert (350 bp) library using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA). The library was sequenced using the Illumina NovaSeq 6000 platform, and the raw data were quality-filtered using the NGS QC Tool Kit v2.3.3 (Patel and Jain Citation2012). High-quality sequence data (4.53 Gb) were then selected for the de novo assembly of the complete chloroplast genome using the assembler SPAdes v3.11.0 (Bankevich et al. Citation2012). Finally, the complete chloroplast genome was annotated using PGA (Qu et al. Citation2019) with the chloroplast genome of Grewia biloba (NC058214) as a reference.
To analyze the G. biloba var. parviflora chloroplast genome, we randomly selected the complete chloroplast genomes of 36 other plant species in the Malvaceae (Yang et al. Citation2018), and two outgroup taxa (Daphne genkwa and Hovenia acerba) were obtained from NCBI. Sequences of 56 common protein-coding genes were obtained from the Grewia biloba var. parviflora. The MAFFT version 7.037 (Katoh and Standley Citation2013) was used to compare the Grewia biloba var. parviflora chloroplast genome with the FFT-NS-2 strategy and 37 other complete chloroplast genomes. A phylogenetic tree of the 38 chloroplast genomes was then constructed using IQ-TREE-1.6.12, based on the maximum-likelihood method with 1000 bootstrap replications and the JTT + F+R3 model, which was selected using ModelFinder (Kalyaanamoorthy et al. Citation2017).
Results and discussion
The chloroplast genome of G. biloba var. parviflora is 158,043 bp in length, with an average sequencing depth of 5420X, and encodes 129 genes; of these, 84 protein-coding genes, eight rRNA genes, and 37 tRNAs. The GC content was 37.4%. The genome includes a large single-copy (LSC) region of 86,957 bp, a small single-copy (SSC) region of 20,138 bp, and two inverted repeat regions (IRs) of 25,474 bp long. In addition, we found that 16 genes (trnK-UUU, rps16, trnG-GCC, atpF, rpoC1, trnL-UAA, trnV-UAC, clpP, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA) all found to contain an intron. The ycf3 gene contains two introns, and the rps12 gene has a trans-splicing condition (). The chloroplast genome of G. biloba var. parviflora was correctly assembled according to the coverage depth (high coverage of over ×100) (Figure S1) (Li et al. Citation2009; Li Citation2013). The maps of the annotated chloroplast genome, cis-splicing genes, and trans-splicing genes of G. biloba var. parviflora (, , respectively) were processed by CPGview (Liu et al. Citation2023).
Figure 2. Circular maps of the chloroplast genome of G. biloba var. parviflora visualized using the CPGview tool (Liu et al. Citation2023). The forward and reverse repeats from the center outward were shown in the first track and connected with green and red arcs, respectively. The tandem repeats and microsatellite sequences were shown in the second and third tracks with different colors. Genes are colored in the outer track by their functional classification listed in the bottom left.
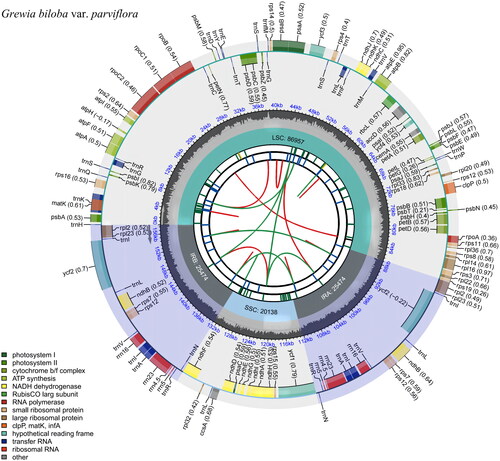
Figure 3. Maximum-likelihood (ML) phylogenetic tree of G. biloba var. parviflora and 37 other species with (A) evolutionary distances and (B) no evolutionary distances. The numbers above the branches indicate the bootstrap support value. The following sequences were used: Gossypium arboretum HQ325740 (Xu et al. Citation2013), Gossypium hirsutum NC007944 (Lee et al. Citation2006), Gossypium herbaceum JF317353, Gossypium aridum NC033396, Gossypium incanum JN019792, Thespesia populnea NC048518, Hibiscus mutabilis MK820657 (Abdullah et al. Citation2020), Abelmoschus manihot NC053353 (Li et al. Citation2020), Malvaviscus penduliflorus NC066439, Hibiscus coccineus OK336487 (Wang et al. Citation2022), Hibiscus sabdariffa MZ522720, Thespesia populnea NC048518, Urena procumbens NC054171 (Wang et al. Citation2021), Talipariti tiliaceum NC053627, Hibiscus sinosyriacus MZ367751, Hibiscus syriacus OM731671, Hibiscus rosa-sinensis NC042239 (Abdullah et al. Citation2020), Malvastrum coromandelianum MK860037 ((Abdullah et al. Citation2020), Sida acuta NC064374, Malva verticillata MT083899 (Wang et al. Citation2020), Alcea rosea NC053839 (Qian et al. Citation2020), Sida szechuensis NC051877, Abutilon theophrasti NC053702 (Lv et al. Citation2021), Tilia amurensis MH169579 (Yang et al.Citation2018), Tilia mongolica NC057237, Tilia endochrysea OK624380, Craigia yunnanensis MN579507, Diplodiscus trichospermus NC065808, Heritiera parvifolia NC038057 (Xin et al. Citation2018), Firmiana simplex NC041438, Theobroma grandiflorum NC054233 (Niu et al. Citation2019), Grewia biloba NC058214 (Xu et al. Citation2021), Colona floribunda NC054164 (Wang et al. Citation2021), Corchorus capsularis NC044467, Corchorus olitorius NC044468, Daphne genkwa MT754180, and Hovenia acerba NC052853 (Zhang et al. Citation2020).
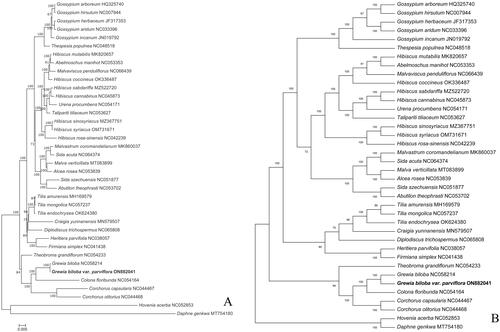
Phylogenetic analysis confirms that G. biloba var. parviflora is on the same branch as the G. biloba, and the genus Grewia and the other genus are on the different branches. The establishment of the phylogenetic tree provides a basis for future research on Malvaceae family ().
Conclusions
The chloroplast genome of Grewia biloba var. parviflora (Bunge) Hand.-Mazz. was sequenced and annotated for the first time. Research shows that the subtle differences in morphology of the G. biloba var. parviflora confirm the genome-wide differences in chloroplasts with the G. biloba, indicating the stability of the variety. It differs from previous studies in that it demonstrates the genetic relationship between G. biloba var. parviflora and G. biloba, providing valuable new genetic information for the genus Grewia. The study through the phylogenetic tree shows the relationships between G. biloba var. parviflora and other species, allowing more accurate plant classification. It provides very valuable data for future in-depth studies of the genus Grewia.
Ethical approval
G. biloba var. parviflora does not belong to the national key protected wild plants category, and collecting it does not violate the Regulations of the People’s Republic of China on Wild Plants Protection. According to Article 5 of the regulation, the state encourages and supports scientific research on wild plants and on-site and ex situ protection of wild plants. No ethical approval or specific permission was needed in this research.
Author contributions
T. G. K. conceived the idea of the study; Y. Y. S. analyzed the data and modified the article; C. B. and L. X. collected and prepared samples; W. J. H. wrote the paper; Y. Y. Y. approved the manuscript to be published; W. X. M. was involved in validation and supervision; all authors discussed the results and revised the manuscript.
Supplemental Material
Download JPEG Image (178.1 KB)Supplemental Material
Download JPEG Image (134.2 KB)Supplemental Material
Download JPEG Image (213.1 KB)Supplemental Material
Download JPEG Image (215.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s). Wen-Juan Hou and Wen-Xiao Men contributed equally to this research. It is worth noting that Wen-Juan Hou and Men Wen-Xiao are co-first authors.
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession no. ON882041. The associated BioProject, SRA, and BioSample numbers are PRJNA852377, SRR19905789, and SAMN29328407, respectively.
Additional information
Funding
References
- Abdullah, Mehmood F, Shahzadi I, Waseem S, Mirza B, Ahmed I, Waheed MT. 2020. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): comparative analyses and identification of mutational hotspots. Genomics. 112(1):581–591. doi: 10.1016/j.ygeno.2019.04.010.
- Abdullah, Mehmood F, Shahzadi I, Ali Z, Islam M, Naeem M, Mirza B, Lockhart PJ, Ahmed I, Waheed MT. 2020. Correlations among oligonucleotide repeats, nucleotide substitutions and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. J Syst Evol. 59(2):388–402. doi: 10.1111/jse.12585.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Chen WS. 1989. The medicinal value of the Grewia biloba. Forest Chem Commun. 5:53.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Lee S-B, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. 2006. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 7:61. doi: 10.1186/1471-2164-7-61.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr arXiv. 3.
- Li HW, Hedge IC. 1933. Malvaceae. In: Symb. Sin, editor. Flora of China. Vol. 17. Beijing, China/St. Louis (MO): Science Press/Missouri Botanical Garden Press; p. 612.
- Li J, Ye G-Y, Liu H-L, Wang Z-H. 2020. Complete chloroplast genomes of three important species, Abelmoschus moschatus, A. manihot and A. sagittifolius: genome structures, mutational hotspots, comparative and phylogenetic analysis in Malvaceae. PLOS One. 15(11):e0242591. doi: 10.1371/journal.pone.0242591.
- Liu JQ, Wu J, Zhang R. 2009. Study on the in vitro antibacterial activity of the chemical constituents of Lentinus edulis. J Jiangxi Coll Tradit Chin Med. 21(2):75–76.
- Liu JQ, Wu JM, Zhang R. 2009. Study on the in vitro antibacterial activities of compounds isolated from Grewia biloba. J Jiangxi Univ Tradit Chin Med. 21(2):75–76.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Lv Y, Yi Y, Han R, Tong X, Takahashi H. 2021. The complete chloroplast genome of Abutilon theophrasti medic (Malvaceae). Mitochondrial DNA B Resour. 6(3):912–913. doi: 10.1080/23802359.2021.1886886.
- Niu YF, Ni SB, Liu J. 2019. The complete chloroplast genome of Theobroma grandiflorum, an important tropical crop. Mitochondrial DNA B Resour. 4(2):4157–4158. doi: 10.1080/23802359.2019.1693291.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619. doi: 10.1371/journal.pone.0030619.
- Qian J, Du Z, Jiang Y, Duan B. 2020. The complete chloroplast genome sequence of Althaea rosea (L.) Cavan. (Malvaceae) and its phylogenetic analysis. Mitochondrial DNA Part B. 5(2):1433–1434. doi: 10.1080/23802359.2020.1736964.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Shi GR, Cheng XL, Liu L, Ma CC. 2006. Plasticity in the anatomical and water physiological characteristics of leaves and secondary xylem of Phyllostachys sylvestris. J Appl Ecol. 10:1801–1806.
- Sun SJ, Meng P, Zhang JS, Jia C, Ren Y. 2014. Ecosystems water use patterns of Quercus variabilis and Grewia biloba based on stable hydrogen and oxygen isotopes in the south aspect of Taihang Mountains. J Ecol. 34(21):6317–6325.
- Wang J-H, Moore MJ, Wang H, Zhu Z-X, Wang H-F. 2021. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene. 765:145103. doi: 10.1016/j.gene.2020.145103.
- Wang L, Cai B, Li J, Yang Y. 2020. The complete chloroplast genome sequence of Malva verticillata. Mitochondrial DNA B. 5(2):1669–1670. doi: 10.1080/23802359.2020.1742602.
- Wang Z, Yang H, Zhang F, Yin Y, Gu C. 2022. The complete chloroplast genome sequence of Hibiscus coccineus. Mitochondrial DNA B Resour. 7(1):217–218. doi: 10.1080/23802359.2021.2022545.
- Xie ZW. 2004. Hanlaying dictionary of Chinese herbal medicine. Beijing: Beijing Science and Technology Press; p. 270.
- Xin G-L, Ren X-L, Liu W-Z, Jia G-L, Deng C-Y. 2018. The complete chloroplast genome of a rare species Heritiera parvifolia Merr. (Malvales: Sterculiaceae). Conserv Genet Resour. 10(4):885–888. doi: 10.1007/s12686-017-0888-9.
- Xu D, Liu M, Ren H, Zhang B, Liu Z, Wang H. 2021. Chloroplast genome and phylogenetic analysis of Grewia biloba. Mitochondrial DNA B Resour. 6(6):1775–1776. doi: 10.1080/23802359.2021.1932626.
- Xu Q, Xiong G, Li P, He F, Huang Y, Wang K, Li Z, Hua J. 2013. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLOS One. 8(5):e37128. doi: 10.1371/annotation/47563c17-536c-465d-9b93-cd35a78f6e66.
- Yang JY, Lee W, Pak J-H, Kim S-C. 2018. Complete chloroplast genome of Ulleung Island endemic basswood, Tilia insularis (Malvaceae), in Korea. Mitochondrial DNA B Resour. 3(2):605–606. doi: 10.1080/23802359.2018.1473731.
- Zhang L, Mao R, Bi H, Shen J, Wang Y, Li M. 2020. Characterization of the complete chloroplast genome of Hovenia acerba (Rhamnaceae). Mitochondrial DNA B Resour. 5(1):934–935. doi: 10.1080/23802359.2020.1714492.
- Zhang XH. 2021. Reproductive ecological characteristics of three pioneer shrubs on Yantai slopes. Shandong: Ludong University.
