Abstract
Macrhybopsis tetranema and Oncorhynchus gilae are fish species endemic to the Southwestern United States. We present the complete mitochondrial genomes for these species. Each genome consisted of 13 protein-coding genes, two ribosomal (rRNA) genes, 22 transfer RNA (tRNA) genes, and the control region (D-loop). Mitogenome lengths were 16,916 base pairs (bp) for M. tetranema, and 16,976 bp for O. gilae. The GC content was 41% for M. tetranema and 46% for O. gilae. The relationships of M. tetranema and O. gilae were consistent with previous phylogenetic analyses.
Introduction
Members of the family Cyprinidae and Salmonidae restricted to the Southwestern United States are threatened by catastrophic events including severe droughts and wildfires. Consequently, numerous species are safe-guarded by captive populations. These populations should reflect standing genetic diversity of the species including protection of genetically distinct lineages such as Evolutionary Significant Units (ESUs) and Management Units (MU) (Moritz Citation1999). For example, mitochondrial sequences from the control region were used to designate Gila Trout (Oncorhynchus gilae Miller 1950) and Apache Trout (O. apache Miller 1972) as distinct ESUs relative to other populations within the Rainbow Trout O. mykiss clade, and to designate O. gilae from the San Francisco River and Gila River drainages as separate MUs (Riddle et al. Citation1998). Subsequently, Wares et al. (Citation2004) identified four unique haplotypes in the remnant San Francisco population. Here, annotated mitochondrial genomes were constructed for the Spruce Creek lineage (San Francisco drainage) of Gila Trout (); a salmonid closely related to Rainbow Trout and for the cyprinid Peppered Chub () (Macrhybopsis tetranema Gilbert et al. Citation2017). These species are listed as threatened (US Fish and Wildlife Service Citation2006) and endangered, respectively (US Fish and Wildlife Service Citation2020). Macrhybopsis tetranema belongs to the M. aestivalis species complex (Eisenhour Citation2004; Gilbert et al. Citation2017) and is now restricted to a 218-km reach of the South Canadian River in New Mexico (NM) and Texas (TX). Its restricted distribution and the constant threat of drought have made establishing a refugial captive population of this species a high priority for management. Oncorhynchus gilae is restricted to isolated populations in the Gila and San Francisco river drainages in southern NM and Arizona (AZ). Populations of O. gilae are threatened by wildfires and introgression with the non-native Rainbow Trout and this species is also supported by captive breeding and augmentation.
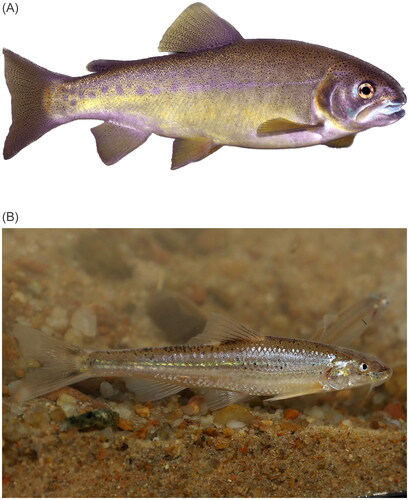
Figure 1. Photographs showing (A) Oncorhynchus gilae (photo credit: Thomas Kennedy, used with permission) and (B) Macrhybopsis tetranema (photo credit: Daniel Fenner, used with permission). Oncorhynchus gilae are characterized by having a stocky, golden-yellowish body, large spots on the adipose fin, numerous fine spots above the lateral line and large spots on the adipose fin (Behnke Citation2002).
Materials
A M. tetranema individual was collected from the South Canadian River, NM (approximate geographic coordinates: 35.389376, −103.355331) in August 2021. Macryhybopsis tetranema is the only species of Macryhybopsis found in the NM portion of the Canadian River. The fish was euthanized with an overdose of MS222. A fin clip from a live O. gilae was collected in December 2021 from the Spruce Creek lineage held at the Mora National Fish Hatchery, NM (approximate locality: 35.994948, −105.319576). In both cases, species identity was determined by visual examination of external morphology by experienced personnel from US Fish and Wildlife Service. Remaining tissue and/or DNA isolates were deposited in the Museum of Southwestern Biology (MSB Cat no. 110774.00 for O. gilae; MSB Cat no. 117041 for M. tetranema; https://msb.unm.edu/, contact: E. DeArmon, [email protected]).
Methods
High-molecular-weight genomic DNA was isolated from muscle and fin tissue from M. tetranema (n = 1) and O. gilae (n = 1), respectively, using QIAGEN® genomic-tips and the protocol outlined by the manufacturer. Whole genomes were sequenced using a PacBio Sequel® II machine for O. gilae (3 SMRT® cells) and M. tetranema (1 SMRT® cell) at the UC Davis Genomics Core using PacBio HiFi SMRTbell® library preparation. Genome assemblies (nuclear plus mitochondrial) were initially constructed from raw PacBio reads with the HiFiAsm assembler v.0.18.1-r466 (Cheng et al. Citation2021) using default parameters for both species, except that the number of haplotypes for O. gilae was set to four since the nuclear genome is tetraploid. Mitogenomes of O. gilae Main Diamond lineage (GenBank: MW300334) and M. hyostoma (GenBank: KX139437) were used to search de novo assemblies for mitochondrial sequences using the CoGeBlast tool (https://genomevolution.org/CoGe/CoGeBlast.pl). For M. tetranema we did not find a good mitogenome assembly using this method and thus a new assembly was performed. First, we searched for potential mitochondrial reads only. Raw reads were mapped to the M. hyostoma mitogenome (GenBank: KX139437) with Bowtie2 v.2.4.2 (Langmead and Salzberg Citation2012) using the “local alignment” and “very sensitive” options. Reads that mapped to the M. hyostoma mitogenome were used as input to HiFiAsm disabling “purge level” and “bloom filter” as suggested by HiFiAsm authors for homozygotic/haploid genomes. Macrhybopsis hyostoma mitogenome was then used to confirm the new assembly using the CoGeBlast tool. Annotation of the mitochondrial genomes was performed using the MitoFish webserver v.3.90 (http://mitofish.aori.u-tokyo.ac.jp/; Iwasaki et al. Citation2013, Citation2018; Zhu et al. Citation2023). DNA sequences were aligned using ClustalW (Thompson et al. Citation1994) implemented in MEGAX (Kumar et al. Citation2018, Stecher et al. Citation2020). Nucleotide substitution models were compared using the maximum likelihood method and the models with the lowest BIC (Bayesian Information Criteria) values were subsequently used for phylogenetic tree construction based on nucleotide sequences. For placing O. gilae in phylogenetic context, Tamura-Nei distances (Tamura and Nei Citation1993) were used and Coho salmon (O. kisutch) was included as an outgroup (). For M. tetranema, the GTR + G model was specified and Roundnose Minnow (Dionda episcopa) was included as an outgroup. Phylogenetic trees were constructed using the program PhyML v. 3.0 (http://atgc.lirmm.fr/phyml; Guindon et al. Citation2010). Due to slightly different lengths of the control region between species, only the coding mitochondrial genes were used to construct phylogenetic trees. Bootstrapping (100 replicates) was used to assess support for nodes. Sequence divergence was calculated between the Spruce Creek individual and other O. gilae lineages and between M. tetranema and other species with the genus for which whole mitogenomes were available.
Results
For O. gilae, the mitochondrial genome was assembled from 48 reads with an average length of 17,019.3 base pairs (bp) ranging from 16,687 to 18,236 bp and coverage was 36.8 × (Supplementary Material, ). Mitochondria for this species comprised 13 protein-coding genes, two rRNA genes, 22 tRNA and a control region (). The length of the O. gilae mitogenome was 16,976 bp. The control region was 1,321 bp in length including a 322 bp insertion comprised of three repetitive elements. This length increase is unique to the Spruce Creek lineage. There was minimal sequence divergence (0.07–0.09%) between the Spruce Creek lineage and the other O. gilae lineages. The Spruce Creek lineage was 0.4% divergent from O. apache. Previous research (, Camak et al. Citation2021) indicated that the Spruce Creek lineage of O. gilae was distinct from the Gila drainage lineages.
Figure 2. Mitochondrial genome maps showing 13 protein coding genes, 2 ribosomal RNAs, 22 transfer RNAs and the control region (D-loop) for (A) Oncorhynchus gilae (GenBank: OQ301638) and (B) Macrhybopsis tetranema (GenBank: OQ301637) generated using the program proksee v. 6.0.2 (https://proksee.ca/) using the relative scale option. Plots of GC content and skew used a window size of 500 and reflect GC content/skew scores on a scale of 0 to 1 using a baseline of 0.5. Positive and negative skew are indicated by values above and below the midpoint respectively. Asterisks in the control region indicate the approximate location and number of repetitive elements. The order of mitochondrial genes is conserved between species.
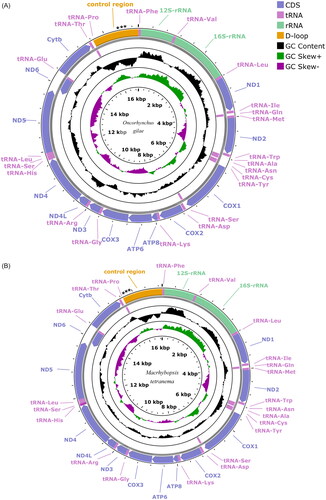
Figure 3. Maximum likelihood trees constructed with the coding sequences of the mitochondrial genomes (A) from Oncorhynchus species including the Spruce Creek lineage of O. gilae (GenBank: OQ301638). Coho salmon (Oncorhynchus kisutch) was included as an outgroup. (B) a subset of cypriniform fishes, including M. tetranema (GenBank: OQ301637) new mitogenome. Roundnose minnow (Dionda episcopa) was included as an outgroup. Node support values were generated with 100 bootstrap replicates. Asterisks and light blue highlighting denote novel sequences presented in this study. GenBank accession numbers are provided near branch tips for all sequences used. References used for comparative analysis are provided in the .
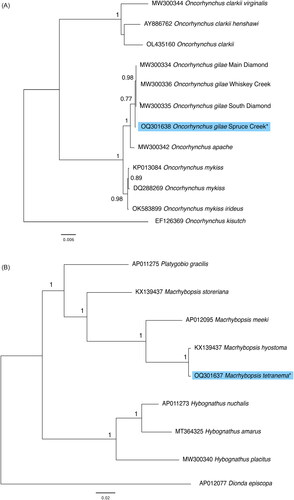
Table 1. List of species, GenBank accession numbers and references for sequences used to construct phylogenetic trees ( and ).
For M. tetranema, the mitochondrial genome was assembled from 51 reads with an average length of 17,045.4 bp ranging from 16,782 to 20,343 bp. Mitogenome coverage was 40 × for M. tetranema. The length of the M. tetranema mitogenome was 16,916 bp and it also comprised 13 protein-coding genes, two rRNA genes, 22 tRNA and a control region (). The control region was 1,256 bp in length and contained a 55 bp repeat. The length of the control region was within the range observed in other species within the genus (1,054 bp [M. storeriana] to 1,264 bp [M. meeki]). Sequence divergence within Macrhybopsis ranged from 0.4% − 10%. Results of the phylogenetic analysis indicate a close relationship between M. tetranema and M. hyostoma while M. meeki is the sister group of this clade as described previously (Echelle et al. Citation2018).
Discussion and conclusions
In this study, mitochondrial genomes were sequenced and assembled for two imperiled species. The order of mitochondrial genes was identical between O. gilae and M. tetranema, members of the Salmonidae and Cyprinidae, respectively. This result is consistent with previous studies showing the mitochondrial gene order is highly conserved among fishes with few exceptions (Miya and Nishida Citation2015). Results presented were consistent with previous research including the distinctness of the Spruce Creek lineage of O. gilae, specifically an increase in the size of the control region caused by repetitive elements in this lineage (Riddle et al. Citation1998). The sequence data presented here could be used to develop a PCR-based diagnostic tool for the identification of Spruce Creek lineage fish. Repeats at the 5′ end on the control region were also identified in M. tetranema while a 74 bp repeat is also present in the control region of M. meeki but not in the basal member of the group (M. storeriana). A close relationship was identified between M. tetranema and M. hyostoma as noted previously and suggesting heterospecific mitochondrial transfer (Echelle et al. Citation2018). The mitochondrial genomes presented here provide an additional resource for ongoing population genetic studies and can be used to inform management efforts for these species.
Ethical approval
This research project has been reviewed and approved by the UNM Institutional Animal Care and Use Committee (IACUC). All investigators in this study have attained animal-use certification regarding the ethical treatment of animals (D16-00565 A4023-01). Sample collection was conducted in accordance with the regulations of the International Union for Conservation of Nature (IUCN).
Authors’ contributions
All authors were involved in study design. MO isolated the DNA of O. gilae and M. tetranema. MO, MBH, and GCD conducted sequence analyses. MO, MBH, GCD and TT drafted and edited the manuscript and approve the study and this manuscript.
Supplemental Material
Download MS Word (182.4 KB)Acknowledgements
Samples were collected with permits from the New Mexico Department of Game and Fish (3015 to TFT and 3764 to Stephen Davenport [US. Fish and Wildlife Service]) and the US. Fish and Wildlife Service (TE38055-0). Samples were collected according to UNM IACUC protocol MSC10-100492-MCC. Images of Gila Trout and Peppered Chub were kindly provided by Thomas Kennedy (UNM) and Daniel Fenner (USFWS) respectively.
Disclosure statement
The authors report no conflicts of interest. The authors are solely responsible for the content and writing of the paper.
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available at NCBI Genbank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession no. OQ301638 for O. gilae and OQ301637 for M. tetranema. The associated sequencing data is available at NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/). For O. gilae the associated BioProject, BioSample and SRA numbers are PRJNA941852, SAMN33611619 and SRR23721617, respectively; for M. tetranema the associated BioProject, BioSample and SRA numbers are PRJNA941280, SAMN33600811 and SRR24116728, respectively.
Additional information
Funding
References
- Behnke R. 2002. Trout and Salmon of North America. New York:Chanticleer Press.
- Brown KH, Drew RE, Weber LA, Thorgaard GH. 2006. Intraspecific variation in the rainbow trout mitochondrial DNA genome. Comp Biochem Physiol Part D Genomics Proteomics. 1(2):219–226. doi: 10.1016/j.cbd.2005.11.004.
- Camak DT, Osborne MJ, Turner TF. 2021. Population genomics and conservation of Gila Trout (Oncorhynchus gilae). Conserv Genet. 22(5):729–743. doi: 10.1007/s10592-021-01355-0.
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18(2):170–175. doi: 10.1038/s41592-020-01056-5.
- Echelle AA, Lang NJ, Borden WC, Schwemm MR, Hoagstrom CW, Eisenhour DJ, Mayden RL, Bussche RA. 2018. Molecular systematics of the North American chub genus Macrhybopsis (Teleostei: Cyprinidae). Zootaxa. 4375(4):537–554. doi: 10.11646/zootaxa.4375.4.4.
- Eisenhour DJ. 2004. Systematics, variation, and speciation of the Macrhybopsis aestivalis complex west of the Mississippi River. Bull Alabama Museum Natural History. 15(23):9–48.
- Gaughan S, Johnson R, Wang J, Wachholtz M, Steffensen K, King T, Lu G. 2016a. The complete mitochondrial genome of the silver chub, Macrhybopsis storeriana. Mitochondrial DNA Part B Resour. 1(1):789–790. doi: 10.1080/23802359.2016.1197053.
- Gaughan S, Johnson R, Wang J, Wachholtz M, Steffensen K, King T, Lu G. 2016b. The complete mitochondrial genome of the shoal chub, Macrhybopsis hyostoma. Mitochondrial DNA Part B Resour. 1(1):911–912. doi: 10.1080/23802359.2016.1197069.
- Gilbert CR, Mayden RL, Powers SL. 2017. Morphological and genetic evolution in eastern populations of the Macrhybopsis aestivalis complex (Cypriniformes: Cyprinidae), with the descriptions of four new species. Zootaxa. 4247(5):501–555. doi: 10.11646/zootaxa.4247.5.1.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi: 10.1093/sysbio/syq010.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540. doi: 10.1093/molbev/mst141.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi: 10.1038/nmeth.1923.
- Miya M, Nishida M. 2015. The mitogenomic contributions to molecular phylogenetics and evolution of fishes: a 15-year retrospect. Ichthyol Res. 62(1):29–71. doi: 10.1007/s10228-014-0440-9.
- Moritz C. 1999. Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas. 130(3):217–228. doi: 10.1111/j.1601-5223.1999.00217.x.
- Osborne MJ, Cameron AC, Fitzgerald BP, McKitrick SA, Paulk MR, Turner TF. 2020. The complete mitochondrial genomes of three imperiled cyprinid fishes Bonytail (Gila elegans), Rio Grande Silvery Minnow (Hybognathus amarus) and Loach Minnow (Tiaroga cobitis). Mitochondrial DNA Part B Resour. 5(3):2368–2370. doi: 10.1080/23802359.2020.1774435.
- Riddle BR, Propst DL, Yates TL. 1998. Mitochondrial DNA variation in Gila trout, Oncorhynchus gilae: implications for management of an endangered species. Copeia. 1998(1):31–39. doi: 10.2307/1447699.
- Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W. 2018. MitoFish and MiFish pipeline: A mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Biol Evol. 35(6):1553–1555. doi: 10.1093/molbev/msy074.
- Stecher G, Tamura K, Kumar S. 2020. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 37(4):1237–1239. doi: 10.1093/molbev/msz312.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. doi: 10.1093/nar/22.22.4673.
- US Fish and Wildlife Service. 2006. Reclassification of the Gila trout (Oncorhynchus gilae) from endangered to threatened; special rule for Gila Trout in New Mexico and Arizona. Fed Regist. 71: 40657–40674.
- US Fish and Wildlife Service. 2020. Endangered and threatened wildlife and plants; endangered species status for the Peppered Chub and designation of critical habitat. Fed Regist. 85:77108–77138.
- Wares JP, Alò D, Turner TF. 2004. A genetic perspective on management and recovery of federally endangered trout (Oncorhynchus gilae) in the American Southwest. Can J Fish Aquat Sci. 61(10):1890–1899. doi: 10.1139/f04-124.
- Zhu T, Sato Y, Sado T, Miya M, and Iwasaki W. 2023. MitoFish, MitoAnnotator, and MiFish pipeline: Updates in ten years. Mol Biol Evol. 40:msad035.
