Abstract
Prionospio Malmgren Citation1867 is one of the abundant genera of the family Spionidae Grube, 1850. Despite its rich diversity, information on their complete mitochondrial genome has remained unknown. In this study, we determined the complete mitochondrial genome of a spionid polychaete, Prionospio cf. japonica Okuda Citation1935. The specimen was collected from the fine sand in the intertidal zone of South Korea. The mitogenome consists of 15,267 base pairs, harboring 13 protein-coding genes (PCGs), 22 transfer RNAs, and two ribosomal RNAs. The maximum-likelihood phylogenetic tree based on the 11 PCGs showed that Prionospio cf. japonica grouped with other spionid polychaetes and formed a monophyletic group. Also, the mtDNA of P. cf. japonica was more closely related to that of non-polydorin spionid, Marenzelleria neglecta, than polydorin spionids. The molecular data will be valuable for studying evolutionary relationships among annelids.
1. Introduction
Prionospio Malmgren Citation1867 is one of the most diverse polychaetous annelid groups belonging to the family Spionidae Grube, Citation1850. More than 100 species are currently known worldwide (Read and Fauchald Citation2022). Among them, P. japonica Okuda Citation1935 was originally described in the brackish water of lakes on the southern coast of Japan. This species has been reported by several authors in Northeast Asia, including China, Japan, and Korea (Imajima and Hartman Citation1964; Paik Citation1975, Citation1982; Zhou and Li Citation2009; Lee et al. Citation2020). It is morphologically characterized by having four pairs of apinnate branchiae (first branchiae distinctly long), prostomium truncates anteriorly with a small median peak, and hooded hooks with four to five pairs of small teeth above the main fang (Okuda Citation1935). Recently, Lee et al. (Citation2020) determined the mitochondrial and nuclear gene fragments of P. japonica based on the specimens collected from the Yellow Sea, Korea. Later, Abe and Sato‐Okoshi (2021) provided the molecular information of P. japonica collected from Sendai Bay, Japan.
Complete mitogenome sequences provide valuable evolutionary information (Chen et al. Citation2016). To date, the complete mitogenome sequences of five spionid species, Boccardiella hamata (Webster Citation1879) (Lee et al. Citation2021), Marenzelleria neglecta (Sikorski and Bick Citation2004; Gastineau et al. Citation2019), Polydora brevipalpa Zachs, Citation1933, Polydora websteri Hartman in Loosanoff & Engle, Citation1943 (Ye et al. Citation2021), and Polydora hoplura (Claparède Citation1868; Lee and Lee Citation2022) have been reported. In this study, we determined the complete mitogenome sequence of P. cf. japonica based on a Korean specimen.
2. Materials and methods
A single adult Prionospio sample () was collected from fine sand in the intertidal zone at Eurwang-dong, Incheon, South Korea (37°24'20.8"N 126°24'49.4"E) using a 500 μm-mesh sieve. Morphological observations were conducted using a stereomicroscope (Leica, Wetzlar, Germany). Species identification was performed based on morphological characteristics. A voucher specimen was deposited at the National Institute of Biological Resources in South Korea (deposit number: NIBRIV0000876631; Url: https://www.nibr.go.kr; Contact person: Min Seock Do, [email protected]). Mitochondria were isolated from live individuals, using the Qproteome Mitochondria Isolation Kit (Qiagen). Mitochondrial DNA (mtDNA) was extracted from the mitochondria using a LaboPass Tissue Mini (Cosmo GENETECH, Seoul, South Korea). Whole-genome amplification (WGA) of the extracted mtDNA was performed using the REPLI-g Mitochondrial DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The amplified mtDNA was sequenced using Illumina HiSeq 4000 (Macrogen, Seoul, Korea), and assembled using Geneious 8.1.9 (Biomatters, Auckland, New Zealand) and NovoPlasty v. 2.7.1 (Dierckxsens et al. Citation2017). The read coverage depth map, assembled using the Geneious program, is presented in Supplementary Figure 1. The complete mitogenome was annotated using the MITOS Webserver (Bernt et al. Citation2013). The extracted mtDNA was deposited in the DNA collection of the National Institute of Biological Resources in South Korea (deposit number: NIBRGR0000645180). The selection of protein-coding genes (PCGs) was conducted using DAMBE 7.0.35 (Xia 2017). DAMBE 7.0.35 was used to determine whether the molecular data is suitable for phylogenetic analysis. A maximum-likelihood tree was constructed using IQ-TREE with the GTR + F + R4 model with 1,000 ultrafast bootstrap replicates (Kalyaanamoorthy et al. Citation2017; Hoang et al. Citation2018). Errantia was used as an outgroup taxon (Perinereis aibuhitensis). The mitogenome sequence obtained was registered in GenBank (GenBank accession number ON470577).
Figure 1. Image of Prionospio cf. japonica Okuda Citation1935 (sample collected from the Yellow Sea of Korea in January 2021; photo by Geon Hyeok Lee).

3. Results and discussion
3.1. Species identification
The most striking characteristics of the Korean specimen are having four pairs of apinnate branchiae (first branchiae distinctly long), prostomium truncates anteriorly with a small median peak, and hooded hooks with four to five pairs of small teeth above the main fang. In this respect, the Korean specimen agrees well with the original description of P. japonica (Okuda Citation1935). However, the pairwise genetic distance of gene fragments between the materials from Korea and P. japonica from Japan was identical in 18S rDNA but showed 8.1% difference in 16S rDNA. The levels of inter-specific differentiation in 16S rRNA are comparable to those observed among other polychaete species, which usually exceed 5% (Wiklund et al. Citation2009; Álvarez-Campos et al. Citation2017; Meißner et al. Citation2017). This result indicated that these are different species. Nevertheless, no morphological differences were observed between the Korean and Japanese specimens (Okuda Citation1935). Therefore, the authors referred to the Korean samples as P. cf. japonica in this study.
3.2. Genomic analysis results
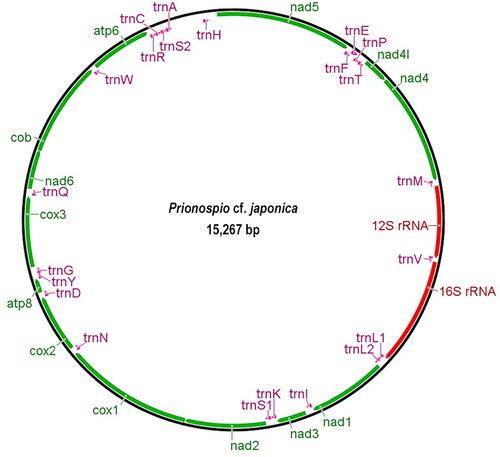
The newly determined complete mitogenome of P. cf. japonica was 15,267 bp in length and contained 13 PCGs, 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and one control region (AT-rich region) (). In the segment between nad6 and nad5, there is a total of 500 bp intergenic spacer that may correspond to the control region. The overall base composition of Korean P. cf. japonica is 32.4% A, 16.2% C, 16.2% G, and 35.3% T, revealing a G + C content of 32.4%.
Figure 2. Mitochondrial genome map of Prionospio cf. japonica Okuda Citation1935.
3.3. Phylogenetic analysis
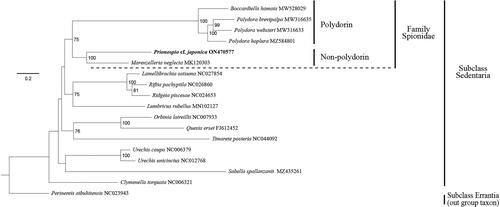
To infer the molecular phylogenetic relationships, a maximum-likelihood tree was constructed based on the concatenated nucleotide sequences of 11 protein-coding genes (cox1, cox2, cox3, cytb, atp6, nad1, nad2, nad3, nad4, nad4l, and nad5) from 16 annelids, including the mtDNA sequence obtained in the present study. The concatenated mtDNA sequences of P. cf. japonica were grouped with those of the other five species of the family Spionidae (Boccardiella hamata, Marenzelleria neglecta, Polydora brevipalpa, Polydora hoplura, and Polydora websteri) (). Prionospio cf. japonica was more closely related to Marenzelleria neglecta, one of the non-polydorin spionids (worms without modified spines in the fifth chaetiger), than to the other four polydorin spionids in the analysis (worms with modified spines in the fifth chaetiger).
4. Conclusion
The assembled mitogenome of P. cf. japonica was 15,267 bp in length and contained 13 PCGs, 22 transfer RNAs, two ribosomal RNAs, and one control region. The phylogenetic analysis of 11 protein-coding genes showed that the sequences of the Korean P. cf. japonica were grouped with those of the other species belonging to the family Spionidae. The newly determined complete mitogenome provides valuable data for further taxonomic and phylogenetic studies.
Ethical approval
This study did not involve endangered or protected species. All the experiments involving animal samples were approved by the animal care and use committee of Inha University (Incheon, South Korea).
Author’s contributions
GH Lee contributed to the sampling in the field, identified worms, performed DNA experiments, analyzed the data, and wrote the manuscript. G-S Min contributed to the design and draft of the manuscript and revising the final version of the manuscript.
All authors agreed to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (41.5 KB)Disclosure statement
The authors declare no conflicts of interest and are responsible for this study.
Data availability statement
The data supporting the findings of this study are openly available in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/ON470577) under accession no. ON470577. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA823822, SRR18651217, and SAMN27361394, respectively.
Additional information
Funding
References
- Álvarez-Campos P, Giribet G, Riesgo A. 2017. The Syllis gracilis species complex: a molecular approach to a difficult taxonomic problem (Annelida, Syllidae). Mol Phylogenet Evol. 109:138–150. doi: 10.1016/j.ympev.2016.12.036.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, P€utz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Chen X, Li M, Liu H, Li B, Guo L, Meng Z, Lin H. 2016. Mitochondrial genome of the polychaete Tylorrhynchus heterochaetus (Phyllodocida, Nereididae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(5):3372–3373. doi: 10.3109/19401736.2015.1018226.
- Claparède É. 1868. Les annélides chétopodes du Golfe de Naples. Mém Soc Phys Hist Nat Genève. 19:313–584.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Gastineau R, Wawrzyniak-Wydrowska B, Lemieux C, Turmel M, Witkowski A. 2019. Complete mitogenome of a Baltic Sea specimen of the non-indigenous polychaete Marenzelleria neglecta. Mitochondrial DNA Part B. 4(1):581–582. doi: 10.1080/23802359.2018.1558125.
- Grube AE. 1850. Die familien der Anneliden. Archiv für Naturgeschichte. 16(1):249–364.
- Hoang DT, Chernomor O, Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281.
- Imajima M, Hartman O. 1964. The polychaetous annelids of Japan. Part II. Allan Hancock Found Publ Occas Pap. 26:1–452.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Lee GH, Lee HE, Min G-S. 2021. The complete mitochondrial genome of Boccardiella hamata (Annelida: Polychaeta: Spionida). Mitochondrial DNA Part B Resour. 6(9):2646–2647. doi: 10.1080/23802359.2021.1964395.
- Lee SJ, Lee SR. 2022. Complete mitochondrial genome of the abalone shell-boring Polydora hoplura (Polychaeta, Spionidae). Mitochondrial DNA Part B Resour. 7(3):438–439. doi: 10.1080/23802359.2022.2047116.
- Lee GH, Yoon SM, Min G-S. 2020. A New Record of Prionospio depauperata (Annelida: Polychaeta: Spionidae) with DNA Barcoding Data of Four Prionospio Species in South Korea. ASED. 36(4):382–386.
- Loosanoff VL, Engle JB. 1943. Polydora in Oysters Suspended in the Water. Biol Bull. 85(1):69–78.
- Malmgren AJ. 1867. Annulata polychæta Spetsbergiæ, Grönlandiæ, Islandiæ et Scandinaviæ hactenus cognita. Öfvers K Vetensk Akad Förh. 24:127–235.
- Meißner K, Bick A, Götting M. 2017. Arctic Pholoe (Polychaeta: Pholoidae): when integrative taxonomy helps to sort out barcodes. Zool J Linn Soc. 179(2):237–262.
- Okuda S. 1935. Some lacustrine polychaetes with a list of brackish-water polychaetes found in Japan. Annot Zool Jpn. 15(2):240–246.
- Paik EI. 1975. The polychaetous annelids in Korea (III). Res Bull Hyosung Women’s Coll. 17:409–438.
- Paik EI. 1982. Taxonomic studies on polychaetous annelids in Korea. Res Bull Hyosung Women’s Coll. 24:745–913. In Korean)
- Read G, Fauchald K. (Ed.) 2022. World Polychaeta Database. World Polychaeta Database. Prionospio Malmgren, 1867. Accessed through: World Register of Marine Species at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=129620 on 2022-02-24.
- Sikorski AV, Bick A. 2004. Revision of Marenzelleria Mesnil, 1896 (Spionidae, Polychaeta). Sarsia. 89(4):253–275. doi: 10.1080/00364820410002460.
- Webster HE. 1879. The Annelida Chaetopoda of the Virginian coast. Trans Albany Inst. 9:202–269.
- Wiklund H, Glover AG, Johannessen PJ, Dahlgren TG. 2009. Cryptic speciation at organic-rich marine habitats: a new bacteriovore annelid from whale-fall and fish farms in the North-East Atlantic. Zool J Linn Soc. 155(4):774–785. doi: 10.1111/j.1096-3642.2008.00469.x.
- Ye L, Yao T, Lu J, Jiang J, Bai C. 2021. Mitochondrial genomes of two Polydora (Spionidae) species provide further evidence that mitochondrial architecture in the Sedentaria (Annelida) is not conserved. Sci Rep. 11(1):13552. doi: 10.1038/s41598-021-92994-3.
- Zachs I. 1933. Annelid worm fauna North-Japanese sea (Polychaeta). K. faune kol´chatykh chervi Severo-Yaponskogo morya (Polychaeta). Gosudarstvennyi Gidrologicheskii Inst, Issled Morei SSSR, Leningrad. 14:125–137.
- Zhou J, Li X. 2009. Report of Prionospio complex (Annelida: Polychaeta: Spionidae) from China’s waters, with description of a new species. Acta Oceanol Sin. 28:116–127.