Abstract
Emberiza godlewskii (Taczanowski, Citation1874) is a passerine bird of eastern Asia which belongs to the genus Emberiza in the bunting family Emberizidae. Obtaining the complete mitochondrial genome sequence of E. godlewskii is helpful to understand the species delimitation for further study of E. cia/godlewskii complex. The circular genome (16,839 bp in length) contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region. The base composition shows that purine percentage (52.87%) is slightly higher than pyrimidine (47.13%). The phylogenetic analysis based on the published data of the mitochondrial genome showed that E. godlewskii is closely related to E. cioides. This new mitochondrial genome data will provide more essential molecular data for further study related to E. cia/godlewskii complex.
Introduction
The species boundary between Emberiza. godlewskii (Taczanowski, Citation1874) and E. cia are more controversial owing to the morphological variation among geographic populations (Vaurie Citation1956; Mauersberger Citation1972; Zheng Citation2005). Our previous research with a small number of gene segments revealed a deep divergence between the southern and northern geographical populations of E. godlewskii (Li et al. Citation2019). Further study on E. cia/godlewskii complex with extensive sampling as well as plumage color and morphological measurements demonstrated that the southern subspecies populations form a distinct monophyletic group, while the northern E. godlewskii is the sister group of Emberiza cia. In other words, there is a distinct southern clade that should be separated from the original E. godlewskii (Li et al. Citation2023). Thus, we recommend the southern E. godlewskii subspecies should be recognized as a full species. However, the full mitochondrial genome of E. godlewskii is still unknown, which limits this species’ ecological and evolutionary research. In this study, we obtained the complete mitochondrial genome sequence of southern E. godlewskii subspecies (E. godlewskii yunnanensis) through high-throughput sequencing for the further study on E. cia/godlewskii complex.
Materials and methods
Ethics statement
The program of sampling collection and experiment in the article has passed the ethical review of Animal and Plant Ethics Committee of Northwest Normal University, and all experiments complied with the guidelines of the committee and the current laws of China.
Sample collection
The genomic DNA was extracted from the muscle tissue of E. godlewskii, which was collected from Jinzhong mountain nature reserve in Guangxi province, China (24.3806˚N 104.5715˚E). A specimen was deposited at the Institute of Zoology and Ecology, College of Life Science, Northwest Normal University (https://sky.nwnu.edu.cn; Dr J. Li, [email protected]) under the voucher number EG2021017.
Mitochondrial genome assembly and annotation
Genomic DNA was isolated using a TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The complete mitochondrial genome of E. godlewskii was sequenced and assembled using Illumina HiSeq 2500 and MitoZ (Meng et al. Citation2019) respectively. The assembled mitochondrial genome was annotated using the MITOS web server (Bernt et al. Citation2013) under the invertebrate mitochondrial code.
Phylogenetic analysis
Sequences used in the phylogenetic analysis were from the NCBI GenBank database and aligned with MAFFT (Katoh and Standley, Citation2013). The phylogenetic analysis was performed using the IQ-TREE 2 (Minh et al. Citation2020)
Results
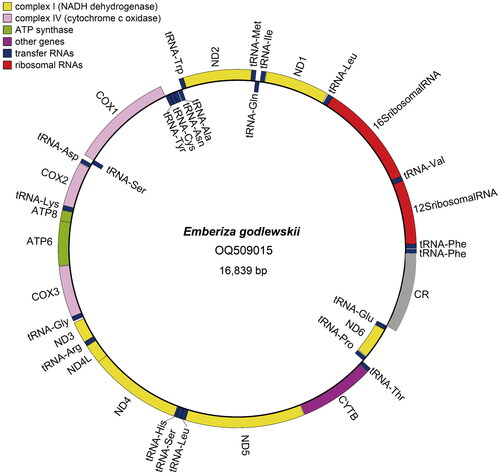
The complete mitochondrial genome of E. godlewskii () is circular and its full length is 16,839 bp. The sequence has been deposited in GenBank under accession number OQ509015. The base composition of mitochondrial DNA (mtDNA) shows that the percentage of A + T (52.87%) is slightly higher than G + C (47.13%). The complete mtDNA sequence has 13 protein-coding genes (PCGs), 23 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and a control region. The 13 PCGs encode ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, CYTB and ND6 respectively. In the13 PCGs, eleven PCGs utilize ATG as the start codon, while COX1 translates from GTG and ND3 from ATA; nine PCGs end with complete (TAA: ND2, COX2, ATP8, ATP6, ND3, ND4L, CYTB; TAG: ND6) or incomplete (T: COX3) stop codons, while COX1 and ND1 terminate with AGG and ND4 and ND5 stop with AGA. The 23 tRNA genes range in size from 66 bp encoding tRNA-Ser to 75 bp encoding tRNA-Leu. The 12S and 16S rRNA genes are 976 bp and 1592 bp respectively. The control region of the mtDNA was 1202 bp in length and distributes between genes of tRNA-Glu and tRNA-Phe ().
Figure 1. The species image of adult E. godlewskii. This bird is characterized by a grey head with chestnut lateral stripe, which is widely distributed in East Asia and tends to select bushy and rocky hill slopes that are often near forests, thickets, ravines, and farm fields. The photograph was taken in Jinzhong mountain nature reserve in Guangxi province of China by Dr X.Bao.

Figure 2. The circular complete mitochondrial genome map of E. godlewskii. The complete mtDNA contains 13 PCGs, 23 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and a control region. The 13 PCGs (ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, CYTB and ND6), 23 tRNA genes, and 2 rRNA genes are encoded in the plus and negative strands of the mtDNA. The control region of the mtDNA distributes between genes of tRNA-Glu and tRNA-Phe. The outside of the ring represents the plus strand, while the inside represents the negative strand.
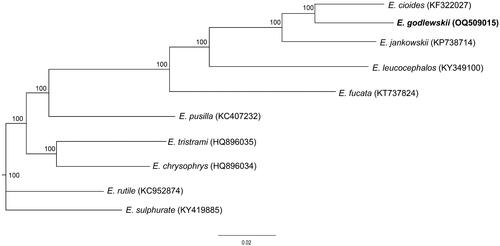
To assess the phylogenetic relationships of E. godlewskii, we selected other nine species of mitochondrial genomes of genus Emberiza birds, which have been published and obtained from GenBank, to construct phylogenetic relationships. We implemented a maximum-likelihood (ML) analysis with the best-fitting model HKY + I + G and 1000 bootstrap replicates. The concatenated nucleotide sequences of 13 PCGs from 10 species of Emberiza genus, which included E. godlewskii, were used to construct the phylogenetic tree. The phylogenetic analysis showed that E. godlewskii is closely related to E. cioides based on these available mitochondrial genome data ().
Figure 3. ML tree showing phylogenetic relationship for ten species of genus Emberiza based on the concatenated nucleotide sequences of 13 PCGs with HKY + I + G model. Numbers above or below nodes indicated the bootstrap support values estimated with 1000 replicates. The following sequences were used: Emberiza cioides, NC_024524, KF322027; E. jankowskii, NC_027251, KP738714; E. leucocephalos, NC_037692, KY349100; E. fucata, NC_033338, KT737824; E. pusilla, NC_021408, KC407232; E. tristrami, NC_015234, HQ896035; E. chrysophrys, NC_015233, HQ896034; E. rutile, NC_024925, KC952874; E. sulfurate, KY419885. The support values are shown next to the nodes. The species under study is bolded.
Discussion and conclusion
In summary, the complete mitochondrial genome of E. godlewskii is 16,839 bp in length, whose size is almost the same as other genus Emberiza birds which have been reported. The phylogenetic relationship based on the published data of mitochondrial genomes demonstrated that E. godlewskii is closely related to E. cioides. However, the phylogenetic position of E. godlewskii is unclear if E. cia mitochondrial genome is added. Thus, the mitochondrial genome of E. godlewskii reported in this study and sequencing the mitochondrial genome of E. cia in the future will provide more essential molecular data for further study related to E. cia/godlewskii complex.
Author contribution
Xin-Kang Bao and Jian-De Li designed the conception of the study; Jian-De Li and Ling-Ling Da performed experiments and data analyses; Jian-De Li drafted the manuscript; Ling-Ling Da revised the manuscript. All authors approved the final manuscript.
Supplemental Material
Download JPEG Image (502.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov under the accession number OQ509015. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA940058, SRR23680762, and SAMN33550910, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Li J, Song G, Liu N, Chang Y, Bao X. 2019. Deep south-north genetic divergence in Godlewski’s bunting (Emberiza godlewskii) related to uplift of the Qinghai-Tibet Plateau and habitat preferences. BMC Evol Biol. 19(1):161. doi: 10.1186/s12862-019-1487-z.
- Li J, Song G, Chen G, Liu N, Dong L, Bao X. 2023. A new bunting species in South China revealed by an integrative taxonomic investigation of the Emberiza godlewskii complex (Aves, Emberidae). Mol Phylogenet Evol. 180:107697. doi: 10.1016/j.ympev.2022.107697.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Mauersberger G. 1972. Über den taxonomischen Rang von Emberiza godlewskii Taczanowski. J Ornithol. 113(1):53–59. doi: 10.1007/BF01647257.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63. doi: 10.1093/nar/gkz173.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Taczanowski L. 1874. Zweiter Nachtrag zum Bericht über die ornithologischen Untersuchungen des Dr. Dybowski in Ost-Sibirien. J Ornithol. 22(3):315–337. doi: 10.1007/BF02004755.
- Vaurie C. 1956. Systematic notes on Palearctic birds. No. 23, Fringillidae: the genera Emberiza, Calcarius, and Plectrophenax. Am Mus Novit. 1805, 1–27.
- Zheng G. 2005. The checklist of the endemic birds of China. A Checklist of the Classification and Distribution of the Birds of China. Science Press: China, Beijing.
