Abstract
Liupao tea is one of the well-known Chinese tea brands and a famous local specialty in Wuzhou, Guangxi, China. However, the genetic background and phylogenetic relationship of the native resource plants of Liupao tea need study, especially at the genomic level. In this study, we reported the complete chloroplast (cp) genome sequence of Camellia sinensis var. sinensis cultivar ‘Liupao’ (LP, Liupao tea population) and inferred its phylogenetic relationship to other tea plant variants or cultivars. The cp genome had a total length of 157,097 bp and the overall GC content was 37.3%. The cp genome contained one LSC region (86,641 bp) and one SSC region (18,276 bp), which were separated by two IR regions (26,090 bp, respectively). Moreover, the cp genomes were composed of 130 genes, including 86 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The phylogenetic analysis showed that LP was closely related to C. sinensis var. pabilimba cv. ‘Lingyunbaihao’. This study will provide useful information for further investigating the genetic background, evolution, and breeding of LP as well as other tea cultivars and varieties.
Introduction
The tea plant species, Camellia sinensis (L.) O. Kuntze 1881, belongs to Theaceae. Camellia sinensis var. sinensis cv. ‘Liupao’ (LP, Liupao tea population) plants, shrubs, or small arbors with bud leaves usually light green or purple, are a breed from natives of Liubao Town, Guangxi, China. LP provides the raw materials used to make Liupao tea, one of the most famous dark tea brands in China, with a history of more than 1500 years. Because of its unique betel nut fragrance, high content of tea polyphenols, aid to digestion as well as durable storage, it is widely favored at home and abroad (Chinese Tea Tree Variety Annals Compilation Committee Citation2001; Ma et al. Citation2016; Qiu et al. Citation2017). With the increase in market demand, the supply of raw materials for LP is insufficient. Some researchers began to study and develop alternative suitable varieties for making Liupao tea (Zhou et al. Citation2013; Zhang et al. Citation2018; Chen et al. Citation2020). Due to the diverse sources of raw materials (varieties) used to make Liupao tea, some Liupao tea products on the market have even lost their original fragrance (Liu et al. Citation2013). Therefore, it is necessary to study the native Liupao tea population, distinguishing it from other tea varieties or cultivars.
High throughput sequencing has dramatically advanced the genetic and genomic studies, including uncovering the phylogeny and evolution of organisms and the molecular regulatory networks underpinning important morphological, agronomic or physiochemical traits. The first tea plant whole genome sequencing (Yunkang 10) was completed in 2017 (Xia et al. Citation2017), which was followed by series of genome sequencing of more tea cultivars and varieties including Shuchazao (Wei et al. Citation2018; Xia et al. Citation2020), Longjin 43 (Wang et al. Citation2020), Biyun (Zhang et al. Citation2020), ancient tea plant DASZ (Zhang et al. Citation2020), Tieguanyin (Zhang et al. Citation2021), Huangdan (Wang et al. Citation2021) and DuyunMaojian (Wang et al. Citation2022). Furthermore, the complete chloroplast (cp) genomes of many more tea plant cultivars, varieties and species in Theaceae have been reported in the recent past (Lee et al. Citation2020; Li et al. Citation2021; Yan et al. Citation2021; Fan et al. Citation2022; Yang et al. Citation2022; Qiao et al. Citation2023). The cp genome sequence data have been widely used in genetics and phylogenetics, resolving enigmas of phylogeny and evolution of numerous taxa from higher taxonomic ranks such as plant kingdom to intraspecific varieties (Li et al. Citation2021). However, neither genome nor cp genome nor else of LP has been sequenced till to now. Studies on genetic diversity and phylogenetic relationship of LP are scarce and only based on SSR or EST-SSR markers (Zhou et al. Citation2011; Huang et al. Citation2021; Wang et al. Citation2022).
Here, we assembled and characterized the complete cp genome of LP and inferred its phylogenetic relationship to other tea varieties, which could contribute valuable information to unveiling genetic diversity, evolution and breeding of LP as well as other tea plants.
Materials and methods
Young leaves (the first leaf under apical bud) of a single LP individual () were collected from Siliu Village, Liubao Town, Cangwu County, Wuzhou City, Guangxi, China (23°49′47” N, 111°19′55” E), and a voucher specimen was deposited at Guangxi Institute of Botany (http://www.gxib.cn/spIBK/, contact person: Chun-Rui Lin, Email: [email protected]) under the voucher number IBK00446204. Total DNA was extracted using the Plant Genomic DNA Kit (TIANGEN, Beijing, China) according to the instructions. Library construction and genomic paired-end (PE150) sequencing was performed on Illumina NovaSeq 6000 instrument (Novogene, Tianjin, China). De novo assemble was constructed using GetOrganelle (Jin et al. Citation2020) with default parameters. The cp genome was annotated using CPGAVAS2 (Shi et al. Citation2019) referring to C. sinensis var. sinensis (NC_020019). The annotated cp genome of LP was submitted to GenBank under the accession number of OQ281601. The cp genome map was drawn using the online software CPGView (http://www.1kmpg.cn/cpgview/). The coverage depth of each base was calculated by mapping all the raw reads to the assembled cp genome using BWA-MEM (Li Citation2013) and SAMtools (Danecek et al. Citation2021), and then ggplot2 (Wickham Citation2016) was used to illustrate the coverage depth map.
Figure 1. The species image of C. sinensis var. sinensis cv. ‘Liupao’. The color of the bud leaves is purple. The photo was taken by the first author.

To unveil the phylogenetic position of LP, we downloaded 37 complete cp genomes of Sect. Thea from NCBI database. The sequences were aligned using MAFFT (Nakamura et al. Citation2018). Then, the maximum-likelihood (ML) implemented in the software IQ-TREE v2 (Minh et al. Citation2020), and the imbed package ModelFinder, was used to reconstruct the phylogenetic tree. The best model detected was GTR + F + I, and the ML tree was built based on this model with 5000 ultrafast bootstraps. Polyspora speciosa was set as the outgroup according to the larger-scale phylogenetic study in Theaceae (Yu et al. Citation2017).
Results
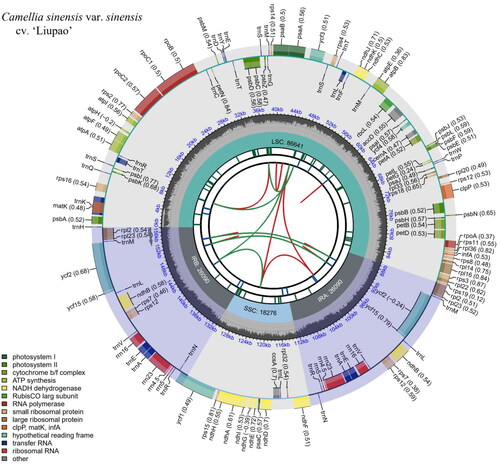
The minimum and average read mapping depth of the assembled genome are 119× and 393× (supplemental Figure S1). Total lengths of the cp genome sequence of LP is 157,097 bp with 37.3% overall GC content (). The cp genome contains one LSC region (86,641 bp) and one SSC region (18,276 bp), which are separated by two IR regions (26,090 bp, respectively). The cp genome contains 130 genes, including 86 protein coding genes, 36 tRNA genes, eight rRNA genes. Among them, 13 cis-splicing genes (supplemental Figure S2) including rps16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, rpl2 (2), ndhB (2), ndhA. The rps12 gene has trans-splicing (supplemental Figure S3), which has three unique exons, and the exon2 and exon3 are duplicated as they are located in the IR regions. The cp genome has 22 genes with intron(s), of which 18 genes have one intron and four genes have two introns.
Figure 2. Schematic map of overall features of the chloroplast genome of C. sinensis var. sinensis cv. ‘Liupao’. The map contains six tracks. From the center outward, the first track shows the dispersed repeats connected by red and green arcs, including direct (D) and palindromic (P) repeats. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the small single-copy (SSC), inverted repeat (Ira and Irb), and large single-copy (LSC) regions. The fifth track plots the GC content along the genome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively.
The phylogenetic tree () shows that LP is closely related to C. sinensis var. pabilimba cv. ‘Lingyunbaihao’ (BS = 85%). These two cp genomes have a total of four bases different, including two mutations and two indels. Previous studies have shown that Lingyunbaihao tea plants can also be used as the raw material to make Liupao tea (Ye et al. Citation2009, Citation2010; Ma et al. Citation2016).
Figure 3. Phylogeny of Camellia sinensis base on the complete chloroplast genome sequences. The cp genome of the accession in bold was sequenced and assembled cp genome in this study. (A) Phylogram tree with a scale bar representing unit of branch length (i.e. the bar length corresponding to substitutions per site). (B) Cladogram tree. Bootstraps support values are shown at the nodes. Three nodes on the phylogenetic tree have no bootstraps support, because IQ-TREE ignores the three sequences: C. sinensis var. sinensis cv. ‘fudingdabaicha’, C. sinensis var. assamica isolate 25D and C. sinensis var. assamica isolate 25E, of which the former is identical to C. sinensis var. assamica cv. ‘duntsa’ and the latter two are identical to C. sinensis var. assamica isolate 25 A. The following published sequences were used: NC_035643 (Yu et al. Citation2017), JQ975032 (Shi et al. Citation2013], NC_039626 (Zeng et al. Citation2018), NC_056149 (Hao et al. Citation2019), KJ806280 (Huang et al. Citation2014), MH460639 (Rawal et al. Citation2020), MH019307 (Zhang et al. Citation2019), MH394408 (Zeng et al. Citation2018), JQ975030 (Shi et al. Citation2013], MH394409 (Zeng et al. Citation2018), MH394410 (Zeng et al. Citation2018), MH394407 (Zeng et al. Citation2018), MT612435 (Li et al. Citation2021), MT773376 (Fan et al. Citation2022), MW046255 (Fan et al. Citation2022), MH042531 (Dong et al. Citation2018), MZ043860 (Yang et al. Citation2022], MZ817088 (Qiao et al. Citation2023), MN086819 (Hao et al. Citation2019), MT773374 (Li et al. Citation2021), MT773375 (Fan et al. Citation2022), KF562708 (Ye et al. Citation2014), MZ153237 (Yan et al. Citation2021), MT773377 (Fan et al. Citation2022), MT773373 (Fan et al. Citation2022), LC488797 (Lee et al. Citation2020], KJ806281 (Huang et al. Citation2014), KJ806279 (Huang et al. Citation2014).
![Figure 3. Phylogeny of Camellia sinensis base on the complete chloroplast genome sequences. The cp genome of the accession in bold was sequenced and assembled cp genome in this study. (A) Phylogram tree with a scale bar representing unit of branch length (i.e. the bar length corresponding to substitutions per site). (B) Cladogram tree. Bootstraps support values are shown at the nodes. Three nodes on the phylogenetic tree have no bootstraps support, because IQ-TREE ignores the three sequences: C. sinensis var. sinensis cv. ‘fudingdabaicha’, C. sinensis var. assamica isolate 25D and C. sinensis var. assamica isolate 25E, of which the former is identical to C. sinensis var. assamica cv. ‘duntsa’ and the latter two are identical to C. sinensis var. assamica isolate 25 A. The following published sequences were used: NC_035643 (Yu et al. Citation2017), JQ975032 (Shi et al. Citation2013], NC_039626 (Zeng et al. Citation2018), NC_056149 (Hao et al. Citation2019), KJ806280 (Huang et al. Citation2014), MH460639 (Rawal et al. Citation2020), MH019307 (Zhang et al. Citation2019), MH394408 (Zeng et al. Citation2018), JQ975030 (Shi et al. Citation2013], MH394409 (Zeng et al. Citation2018), MH394410 (Zeng et al. Citation2018), MH394407 (Zeng et al. Citation2018), MT612435 (Li et al. Citation2021), MT773376 (Fan et al. Citation2022), MW046255 (Fan et al. Citation2022), MH042531 (Dong et al. Citation2018), MZ043860 (Yang et al. Citation2022], MZ817088 (Qiao et al. Citation2023), MN086819 (Hao et al. Citation2019), MT773374 (Li et al. Citation2021), MT773375 (Fan et al. Citation2022), KF562708 (Ye et al. Citation2014), MZ153237 (Yan et al. Citation2021), MT773377 (Fan et al. Citation2022), MT773373 (Fan et al. Citation2022), LC488797 (Lee et al. Citation2020], KJ806281 (Huang et al. Citation2014), KJ806279 (Huang et al. Citation2014).](/cms/asset/1e3ce2bb-d946-4386-bf67-3d0fb4eefe0f/tmdn_a_2250072_f0003_b.jpg)
Discussion and conclusion
This is the first report of the sequence and features of the complete cp genome of LP, and also the first time to reveal the phylogenetic relationship of LP based on sequences. Only a few previous studies have analyzed the genetic relationship between LP and other tea plants but they are only based on the SSR or EST-SSR molecular markers (Zhou et al. Citation2011; Wang et al. Citation2022). The phylogenetic relationship of LP in this study is inconsistent with these two studies, and the closest related variety of LP was not included in these two studies. In this study, LP and C. sinensis var. pabilimba cv. ‘Lingyunbaihao’ was closest, but these two cultivars belong to different varietas in C. sinensis. This is best explained by hybridization introgression between C. sinensis var. pabilimba cv. ‘Lingyunbaihao’ and LP with the former capturing the cp genome of the latter according to the phylogeny that two samples of C. sinensis var. pabilimba are divided into two distantly-related clades, one is outside of all C. sinensis samples, while C. sinensis var. pabilimba cv. ‘Lingyunbaihao’ is embedded in the C. sinensis var. sinensis clade. This hypothesis would be further validated based on nuclear gene data and more samples of LP and C. sinensis var. pabilimba cv. ‘Lingyunbaihao’. Besides, considering the large number of different tea plant cultivars and the few published cp genomes, the phylogeny of LP merits further test with more varieties and cultivars sampled as well. Anyway, this research could benefit future studies on the genetic diversity, evolution and breeding of LP.
Author contributions
Yan-ni Liang: collected materials, prepared for sequencing, and wrote the manuscript. Hong Li and Xi-yang Huang: performed the data analyses. Yue-jing Bin and Yu-mei Zhen: conducted the experiment. Xin-mei Qin: conceived the work and revised the manuscript. All authors have read and agreed to publish the manuscript.
Ethical statement
The samples used in this study were planted by a tea farmer and have received permission for sample collection from the tea farmer.
Supplemental Material
Download MS Word (261.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OQ281601. The associated BioProject, SRA, and BioSample numbers are PRJNA924054, SRR23093613 and SAMN32745220, respectively.
Additional information
Funding
References
- Chinese Tea Tree Variety Annals Compilation Committee. 2001. The variety annals of tea trees in China. Shanghai: Shanghai Science and Technology Publishing House. p. 187.
- Chen J, Liao CS, Liu CM, Teng MY, Qin XJ. 2020. Introduction to Guangxi clonal tea varieties suitable for Liupao tea production. South China Agric. 14(19):53–56. doi: 10.19415/j.cnki.1673-890x.2020.19.014
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience. 10(2):giab008. doi: 10.1093/gigascience/giab008.
- Dong M, Liu SQ, Xu ZG, Hu ZY, Ku WZ, Wu L. 2018. The complete chloroplast genome of an economic plant, Camellia sinensis cultivar Anhua, China. Mitochondrial DNA B Resour. 3(2):558–559. doi: 10.1080/23802359.2018.1462124.
- Fan L, Li L, Hu YF, Huang YB, Hong YC, Zhang B. 2022. Complete chloroplast genomes of five classical Wuyi tea varieties (Camellia sinensis, Synonym: thea bohea L.), the most famous Oolong tea in China. Mitochondrial DNA B Resour. 7(4):655–657. doi: 10.1080/23802359.2022.2062263.
- Hao WJ, Ma JQ, Ma CL, Jin JQ, Chen L. 2019. The complete chloroplast genome sequence of Camellia tachangensis F.C. Zhang (Theaceae). Mitochondrial DNA B Resour. 4(2):3344–3345. doi: 10.1080/23802359.2019.1673247.
- Hao WJ, Wang SL, Yao MZ, Ma JQ, Xu YX, Chen L. 2019. The complete chloroplast genome of an albino tea, Camellia sinensis cultivar ‘Baiye 1’. Mitochondrial DNA B Resour. 4(2):3143–3144. doi: 10.1080/23802359.2019.1667889.
- Huang H, Shi C, Liu Y, Mao S, Gao L. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14(1):151. doi: 10.1186/1471-2148-14-151.
- Huang HC, Jiang WX, Liang CX, Yao X, Nie HQ, Bai TD. 2021. EST-SSR-based analysis on genetic diversity of ancient Liupao tea trees and their progeny in Guangxi. Mol Plant Breed. 19(7):2410–2418.
- Jin JJ, Yu WB, Yang JB, Yu S, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Lee DJ, Kim CK, Lee TH, Lee SJ, Moon DG, Kwon YH, Cho JY. 2020. The complete chloroplast genome sequence of economical standard tea plant, Camellia sinensis L. cultivar Sangmok, in Korea. Mitochondrial DNA B Resour. 5(3):2835–2836. doi: 10.1080/23802359.2020.1790311.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997v1302 [q-bio.GN].
- Li HT, Luo Y, Gan L, Ma PF, Gao LM, Yang JB, Cai J, Gitzendanner MA, Fritsch PW, Zhang T, et al. 2021. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 19(1):232. doi: 10.1186/s12915-021-01166-2.
- Li L, Hu Y, He M, Zhang B, Wu W, Cai PM, Huo D, Hong YC. 2021. Comparative chloroplast genomes of the tea plants and the implications for the different origins of the two Assam teas. BMC Genomics. 22(1):138. doi: 10.1186/s12864-021-07427-2.
- Li L, Hu YF, Wu LH, Chen RB, Luo SC. 2021. The complete chloroplast genome sequence of Camellia sinensis cv. Dahongpao: a most famous variety of Wuyi tea (Synonym: thea bohea L.). Mitochondrial DNA B Resour. 6(1):3–5. doi: 10.1080/23802359.2020.1844093.
- Liu XD, Yang C, Tang ZB, Liu YF. 2013. Research on the genetic marks of tea plants that suitable to make Liubao tea and their roles in the breeding of Liubao tea. Chin Agric Sci Bull. 29(4):136–140.
- Ma SC, Yang M, Long ZR. 2016. Liubaocha Daguan. Guilin: Lijiang Publishing House. p. 245–250.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492. doi: 10.1093/bioinformatics/bty121.
- Qiao DH, Yang C, Guo Y. 2023. The complete chloroplast genome sequence of Camellia sinensis var. sinensis cultivar ‘FuDingDaBaiCha’. Mitochondrial DNA B Resour. 8(1):100–104. doi: 10.1080/23802359.2022.2161327.
- Qiu RJ, Ma SC, Huang LY, Lonag ZR, Yu CP, Cao ZH, Zheng XQ. 2017. Preliminary study on Liupao tea germplasm resources. J Tea. 43(1):28–31.
- Rawal HC, Kumar PM, Bera B, Singh NK, Mondal TK. 2020. Decoding and analysis of organelle genomes of Indian tea (Camellia assamica) for phylogenetic confirmation. Genomics. 112(1):659–668. doi: 10.1016/j.ygeno.2019.04.018.
- Shi C, Liu Y, Huang H, Xi EH, Zhang HB, Gao LZ, Su B. 2013. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PloS One. 8(3):e59620. doi: 10.1371/journal.pone.0059620.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Wang F, Zhang BH, Wen D, Liu R, Yao XZ, Chen Z, Mu R, Pei HM, Liu M, Song BX, et al. 2022. Chromosome-scale genome assembly of Camellia sinensis combined with multi-omics provides insights into its responses to infestation with green leafhoppers. Front Plant Sci. 13:1004387. doi: 10.3389/fpls.2022.1004387.
- Wang LB, Huang LY, Teng CQ, Wu LY, Cheng H, Yu CP, Wang LY. 2022. Genetic and phylogenetic analysis for germplasm resources of Camellia sinensis from Wuzhou city. J Tea Sci. 42(5):601–609.
- Wang PJ, Yu JX, Jin S, Chen S, Yue C, Wang WL, Gao SL, Cao HL, Zheng YC, Gu MY, et al. 2021. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic Res. 8(1):107.
- Wang XC, Feng H, Chang YX, Ma CL, Wang LY, Hao XY, Li A, Cheng H, Wang L, Cui P, et al. 2020. Population sequencing enhances understanding of tea plant evolution. Nat Commun. 11(1):4447. doi: 10.1038/s41467-020-18228-8.
- Wei CL, Yang H, Wang SB, Zhao J, Liu C, Gao LP, Xia EH, Lu Y, Tai YL, She GB, et al. 2018. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A. 115(18):E4151–E4158. doi: 10.1073/pnas.1719622115.
- Wickham H. 2016. Data Analysis. In: Wickham H, editor. Ggplot2: elegant graphics for data analysis. Cham: springer International Publishing. p. 189–201.
- Xia EH, Tong W, Hou Y, An YL, Chen LB, Wu Q, Liu YL, Yu J, Li FD, Li RP, et al. 2020. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol Plant. 13(7):1013–1026. doi: 10.1016/j.molp.2020.04.010.
- Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W, et al. 2017. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 10(6):866–877. doi: 10.1016/j.molp.2017.04.002.
- Yan MH, Liu K, Wang M, Lyu Y, Zhang Q. 2021. Complete chloroplast genome of Camellia sinensis cv. Xinyang 10 and its phylogenetic evolution. J Tea Sci. 41(6):777–788.
- Yang C, Qiao DH, Guo Y, Chen J, Chen ZW. 2022. The complete chloroplast genome sequence of Camellia sinensis cultivar ‘Qiancha1’ from Guizhou Province, China. Mitochondrial DNA B Resour. 7(2):404–405. doi: 10.1080/23802359.2021.2005490.
- Ye JP, He TQ, Jiang MG. 2009. Adaptability test of Liupao Brick tea of Lingyun Baihao tea variety. Guangdong Tea Ind. (6):33–35.
- Ye JP, He TQ, Jiang MG. 2010. Techniques for manufacture of Liupao Brick tea by leaves of Linyun Baihao tea variety. China Tea. 32(8):22–23.
- Ye XQ, Zhao ZH, Zhu QW, Wang YY, Lin ZX, Ye CY, Fan LJ, Xu HR. 2014. Entire chloroplast genome sequence of tea (Camellia sinensis cv. Longjing 43): a molecular phylogenetic analysis. J Zhejiang Univ. 40(4):404–412.
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215(3):1235–1248. doi: 10.1111/nph.14683.
- Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14(:43. doi: 10.1186/s13007-018-0300-0.
- Zhang F, Li W, Gao C, Zhang D, Gao L. 2019. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. Sci Data. 6(1):209. doi: 10.1038/s41597-019-0201-8.
- Zhang F, Wen LX, Peng JR, Huang SH, Tan YW, Huang XX. 2018. Research progress of Guangxi Liupao tea. J Tea Commun. 45(3):13–16.
- Zhang QJ, Li W, Li K, Nan H, Shi C, Zhang Y, Dai ZY, Lin YL, Yang XL, Tong Y, et al. 2020. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol Plant. 13(7):935–938. doi: 10.1016/j.molp.2020.04.009.
- Zhang WY, Zhang YJ, Qiu HJ, Guo YF, Wan HL, Zhang XL, Scossa F, Alseekh S, Zhang QH, Wang P, et al. 2020. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat Commun. 11(1):3719. doi: 10.1038/s41467-020-17498-6.
- Zhang XT, Chen S, Shi LQ, Gong DP, Zhang SC, Zhao Q, Zhan DL, Vasseur L, Wang YB, Yu JX, et al. 2021. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat Genet. 53(8):1250–1259. doi: 10.1038/s41588-021-00895-y.
- Zhou WQ, Nong YF, Su SM, Guo WS, Zhang XY. 2013. On the relationship between the quality of Liupao tea and Liupao population tea plants. Chin for Food Ind. (6):34–35.
- Zhou YH, Qiao XY, Ma CL, Qiao TT, Jin JQ, Yao MZ, Chen L. 2011. Genetic diversity and structure of tea landraces from Guangxi based on EST-SSR analysis. Sci Silvae Sin. 47(3):59–67.
