Abstract
Heritiera fomes Buch.-Ham. (1800) is a species of mangrove in the family Malvaceae, widely distributed in the Indo-Pacific and listed as ‘endangered’ (EN) on the International Union for Conservation of Nature’s (IUCN) red list. We reported the complete chloroplast genome sequence of H. fomes. The genome was 168,521 bp in length and included two inverted repeats (IRs) of 34,496 bp, separated by a large single-copy (LSC) region of 88,604 bp and a small single-copy (SSC) region of 10,925 bp, respectively. The genome contained 87 protein-coding genes (PCGs), 8 rRNA genes, and 37 tRNA genes. The maximum-likelihood (ML) phylogenetic tree suggested that H. fomes is closely related to Heritiera angustata and Heritiera parvifolia with relatively high support bootstrap values of 86% and 100% with other species (Heritiera littoralis and Heritiera javanica), suggesting a relatively close genetic relationship between the five Heritiera plants. The chloroplast genome sequence provided a useful resource for conservation genetics studies of H. fomes and for phylogenetic studies of Heritiera.
Introduction
Heritiera fomes Buch.-Ham. (1800) is a species of mangrove in the family Malvaceae. Its common names are Sander, Sandri, Jekanazo, and Pinlekanazo. It is native to the Indo-Pacific coasts of Bangladesh, Malaysia, Myanmar, and Thailand (Kathiresan et al. Citation2010). H. fomes is currently listed as an endangered (EN) species on the International Union for Conservation of Nature’s (IUCN) Red List (https://www.iucnredlist.org/species/178815/7615342) as the population of this mangrove species is declining due to human activities such as aquaculture, agriculture, and coastal land development, resulting in a rapid decline of mangrove populations (Kathiresan et al. Citation2010).
Recently, the complete chloroplast genomes of several Heritiera species, such as Heritiera angustata (Zhao et al. Citation2018), Heritiera parvifolia Merr (Xin et al. Citation2018), and Heritiera javanica (Zhang et al. Citation2020), have been sequenced. Here, we report the complete chloroplast genome of H. fomes and the phylogenetic tree in the Heritiera genus. The chloroplast genome of H. fomes provides basic genetic data, that will be helpful for understanding its characteristics, phylogenetics and evolution. It will be useful for conservation, genetic studies within the genus Heritiera, and phylogenetic studies of Heritiera.
Materials
Fresh leaves of H. fomes were identified by Suchart Yamprasai ([email protected]) and collected from the Mangrove Forest Research Center, Ranong, Thailand (10°10′16.7″N, 98°42′27.9″E), following the guidelines on the implementation of the ‘IUCN Policy Statement on Research Involving Species at Risk of Extinction,’ with special reference to the Scientific Collecting of Threatened Species (July 2022). The sample collection for this study was permitted by the Department of Marine and Coastal Resources (Ministry of Natural Resources and Environment, Thailand) under project number P-1952261. The herbarium voucher specimen and genomic DNA of H. fomes were deposited at the National Biobank of Thailand (NBT), National Science and Technology Development Agency (NSTDA) (https://www.nationalbiobank.in.th/en/plant-services; contacts: Anuttara Nathalang, [email protected] and Panyavut Aumpuchin, [email protected]) under the voucher numbers BBH004068 and NBTG000003, respectively. H. fomes had the following morphologic characteristics in this study: 14 m height, grayish-brown smooth bark, and smooth leaves. The fruit carpels on the pink or orange bell-shaped flowers can measure up to 5 and 5 mm in diameter ().
Methods
Genomic DNA from H. fomes leaves was extracted using the MagAttract HMW DNA Kit (Qiagen, Germany) and then used for genomic library preparation. Paired-end reads (150 bp) were sequenced using the HiSeq X Ten platform (Illumina, San Diego, CA). The H. fomes chloroplast genome was assembled using Get Organelle (Jin et al. Citation2020) with a coverage of 3788× (Figure S1). The chloroplast genome was annotated using GeSeq (Tillich et al. Citation2017) and manually corrected for start and stop codons as well as for intron-exon after comparison with reported H. fomes chloroplast genes. All transfer RNAs (tRNAs) were predicted using ARAGORN version 1.2.36 (Laslett and Canback, Citation2004) implemented in the GeSeq software. The complete chloroplast genome of H. fomes was submitted to the GenBank database of the National Center for Biotechnology Information (NCBI) under accession number OM022247. The circular genome map was drawn using the CPGView program (http://www.1kmpg.cn/cpgview/; Liu et al. Citation2023).
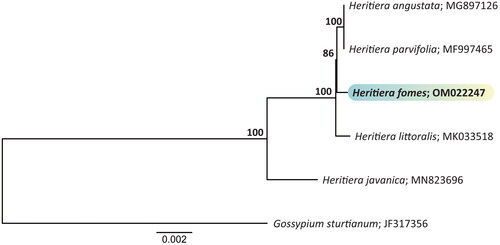
In phylogenetic analysis, to understand the phylogenetic position of H. fomes within the Heritiera genus. All five chloroplast genome sequences in the Heritiera genus (family Malvaceae) are available in GenBank (H. angustata, H. javanica, H. littoralis, H. parvifolia, and H. fomes), using Gossypium sturtianum as an outgroup. We identified orthologous groups from 1348 proteins in the chloroplast genome sequences of H. fomes and five plant species using OrthoFinder software (Emms and Kelly Citation2019). Then, 63 conserved chloroplast protein-coding genes (PCGs) were constructed into a maximum-likelihood (ML) phylogenetic tree using the RAxML-ng tool (Kozlov et al. Citation2019) with 1000 bootstrap replicates. The ModelTest-NG program (Darriba et al. Citation2020) was used to estimate the substitution models for the ML tree construction.
Results
The full-length complete chloroplast genome of H. fomes was 168,521 bp long. Its circular, double-stranded molecule contained two inverted repeats (IRs) of 34,496 bp, separated by a large single-copy (LSC) region and a small single-copy (SSC) region of 88,604 and 10,925 bp, respectively (). The overall GC content of the H. fomes chloroplast genome was 36.88%, while those of the LSC, SSC, and IR regions were 35.04%, 31.73%, and 40.06%, respectively. A total of 132 genes were identified in the chloroplast genome of H. fomes, including 87 PCGs, 8 ribosomal RNA (rRNA) genes, and 37 tRNA genes. It included 15 Cis-splicing genes (atpF, clpP, petB, petD, rpl16, rpoC1, trnI, trnK-UUU, trnL-UAA, trnS-CGA and ycf3; ndhB, rpl2, trnA-UG and trnE-UUC. Four of these genes (ndhB, rpl2, trnA-UG, and trnE-UUC) were duplicated and one appeared as a trans-spliced gene (rps12). Two of the Cis-splicing genes (ycf3 and clpP) contained two introns, the other genes contained one intron. The structures of the 12 protein-coding trans- and cis-splicing genes are shown in Figure S2. The ML phylogenetic tree confirmed that the plastome genome of the target species is closely related to the other Heritiera plastomes ().
Figure 2. Genetic map of the complete chloroplast genome of H. fomes. The map contains six circles. From the center going outward, the first circle shows the distributed repeats connected with red (the forward direction) and green (the reverse direction) arcs. The next circle shows the long tandem repeats marked with short blue bars. The third circle shows the short tandem repeats (STRs) or microsatellite sequences as short bars with different colors. The fourth circle shows the size of the LSC, SSC, IRA, and IRB. The fifth circle shows the GC contents along the plastomes. The sixth circle shows the genes, and their optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
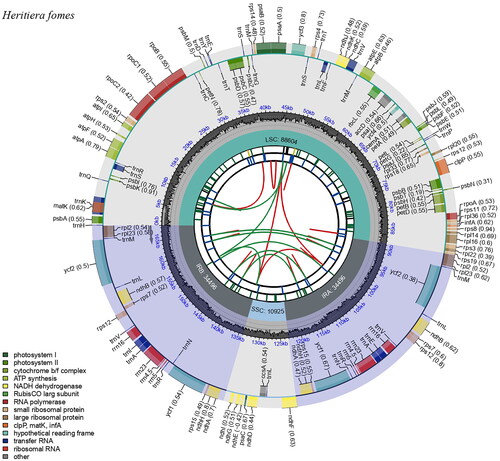
Figure 3. The phylogenetic tree shows the taxonomic relationships of H. fomes and four plastomes in the Heritiera genus, using Gossypium sturtianum as an outgroup. The numbers on the internal nodes were bootstrap values from 1,000 replicates. H. fomes (OM022247) is the target chloroplast sequence of this study, highlighted with color. The following sequences were used: Gossypium sturtianum JF317356 (Unpublished), H. angustata MG897126 (Zhao et al. Citation2018), Heritiera fomes OM022247 (this study), H. javanica MN823696 (Zhang et al. Citation2020), H. littoralis MK033518 (Unpublished), and H. parvifolia MF997465 (Xin et al. Citation2018).
Discussion and conclusions
The complete chloroplast genome of H. fomes reported in this study provided basic genetic data and showed the phylogenetic relationships with four Heritiera plastomes to confirm that the plastome genome of the target species is closely related to the other Heritiera plastomes. These results will provide a useful resource for conservation, genetics and phylogenetic studies of Heritiera species.
Author contributions
TY, WP, and ST designed the research study and obtained the funding. SY, CN, and PP performed laboratory work (sample collection, DNA extraction, library construction, and sequencing). WK and CS performed bioinformatics analyses. TY wrote and revised the manuscript, and all authors reviewed it.
Ethical approval
Research and collection of plant material were conducted according to the guidelines for the implementation of the ‘IUCN Policy Statement on Research Involving Species at Risk of Extinction,’ with special reference to the Scientific Collecting of Threatened Species (July 2022) provided by the Mangrove Forest Research Center, the Department of Marine and Coastal Resources. Permission was granted by the Department of Marine and Coastal Resources (Ministry of Natural Resources and Environment, Thailand) under grant number P19-52261 to carry out research on the species.
Supplemental Material
Download JPEG Image (138.9 KB)Supplemental Material
Download JPEG Image (3.3 MB)Supplemental Material
Download JPEG Image (2.1 MB)Supplemental Material
Download MS Word (1.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OM022247. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA783468, SRR17659486, and SAMN23430239, respectively.
Additional information
Funding
References
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. doi: 10.1093/molbev/msz189.
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. doi: 10.1186/s13059-019-1832-y.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Kathiresan K, Salmo SG, Fernando ES, Peras JR, Sukardjo S, Miyagi T, Ellison J, Koedam NE, Wang Y, Primavera J, et al. 2010. Heritiera fomes. The IUCN red list of threatened species doi: http://dx.doi.org/10.2305/IUCN.UK.20102.RLTS.T178815A7615342.en.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi: 10.1093/bioinformatics/btz305.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. doi: 10.1093/nar/gkh152.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView, a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Xin GL, Ren XL, Liu WZ, Jia GL, Deng CY. 2018. The complete chloroplast genome of a rare species Heritiera parvifolia Merr. (Malvales: Sterculiaceae). Conservation Genet Resour. 10(4):885–888. doi: 10.1007/s12686-017-0888-9.
- Zhang J, Li Y, Heng S, Wang Y. 2020. The complete chloroplast genome sequence of Heritiera javanica. Mitochondrial DNA Part B. 5(2):1174–1175. doi: 10.1080/23802359.2020.1731346.
- Zhao KK, Wang JH, Cai YC, Zhu ZX, Lopez-Pujol J, Wang HF. 2018. Complete chloroplast genome sequence of Heritiera angustata (Malvaceae): an endangered plant species. Mitochondrial DNA B Resour. 3(1):141–142. doi: 10.1080/23802359.2017.1422398.

