Abstract
Anabarilius duoyiheensis is a native and rare fish in Yunnan. In this study, the complete mitochondrial genome of A. duoyiheensis was sequenced and published for a total of 16,614 bp, including 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and one control region. The phylogenetic analysis based on the complete mitochondrial genome showed that A. duoyiheensis belongs to the clade of the genus Anabarilius and was sister to the clade of Hemiculter. This study also contributes to the genus phylogeny of Anabarilius and other members of the family Xenocyprididae.
1. Introduction
Anabarilius duoyiheensis Li, Mao & Lu, Citation2002 belongs to Cypriniformes, Xenocyprididae, Anabarilius and distributed in Duoyi river in Qujing, Yunnan, China (Li et al. Citation2002; Chen X Citation2013). The morphological features of A. duoyiheensis were: mouth terminal; head length is significantly greater than body height; arcuate abdomen; incomplete ventral edge; long pectoral fins with terminal tip; short branchial rake (Li et al. Citation2002). Some of the main features were similar to those of A. macrolepis with distinguishable features: fewer branchial rake (7–8); the end of unbranched dorsal fin is hard; longer pectoral fin (Li et al. Citation2002). The Anabarilius species, distributed mainly in central and eastern Yunnan and south of Sichuan Province, were peculiar to China (Yang J and Chu Citation1987). However, there are no data regarding A. duoyiheensis and particularly its mitochondrial genome. Here, we presented the mitochondrial genome of A. duoyiheensis by using next-generation sequencing, and these results will be useful in further phylogenetic analysis of genus of Anabarilius.
2. Materials and methods
2.1. Specimens collection and preservation
The biological specimens of A. duoyiheensis were sampled from the Lu Buge reach of the Duoyi river (N: 24°45.669′, E: 104°29.758′) in Qujing, Yunnan, and the live fish were kept in Lubuge Township Fisheries Station. A specimen was deposited at Yunnan Institute of Fishery Sciences Research with the specimen voucher number: DYBY-01 (contact person: Junjie Wu, [email protected]). The specimen of A. duoyiheensis was identified by morphological feature according to Li’s description (Li et al. Citation2002), and descriptive image was taken by Leng (corresponding author) ().
2.2. Illumina sequencing, assembly, and annotation
The tissue samples were homogenized and DNA was extracted by following ammonium acetate precipitation method (Rivero et al. Citation2006). The Library was prepared according to Illumina protocol, and sequenced in Illumina NovaSeq platform (Modi et al. Citation2021). The contigs were assembled using SPAdes V3.13 with -k 127 (Bankevich et al. Citation2012). The functional annotation of mitochondrial genome was performed using MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Donath et al. Citation2019).
2.3. Phylogenetic analysis
To analyze the phylogenetic relationship of A. duoyiheensis, published mitochondrial sequences of other 18 species in the family of Xenocyprididae were chosen; the species information and references are listed in . The alignment sequences were obtained in MEGA X using MUSCL method (Kumar et al. Citation2008), and substitution model was evaluated in jModeltest 2.1.3 (Posada Citation2008). The AICc results were carried in phyML 3.0 for rebuilding the maximum-likelihood (ML) tree (Guindon et al. Citation2009). S. curriculus (Zeng S et al. Citation2016) was chosen and regarded as outgroup.
Table 1. Whole mitochondrial genomes used to infer the phylogenetic analysis.
3. Results
3.1. Mitochondrial characterization
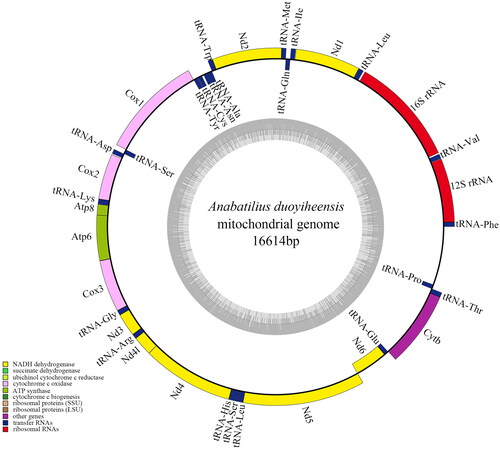
The complete mitochondrial DNA sequence of A. duoyiheensis was 16,614 bp in length with a depth of 582.42x (Figure S1, Table S1), including 13 protein-coding genes, 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a D-loop control region (). The initiation codon of 13 protein coding genes began with ATG except COX1, which began with GTG. Incomplete stop codons were identified in Nd2, Cox2, Cox3, Nd3, Nd6, and Cytb; TAA or TAG stop codon was identified in Nd1, Cox1, Atp8, Atp6, Nd4l, and Nd5. The base composition was 28.87%A, 24.89%T, 27.86%C, and 18.37%G, the A + T content was 53.76% and G + C content was 46.24%. Most of the genes were in the heavy strand (H-strand), except for ND6 and eight tRNAs. The blast search shows that the identity of A. duoyiheensis mitochondrial sequences was 94% similar to A. liui yalongensis, 93% to A. brevianalis, and 91% to H. leucisculus and H. tchangi.
3.2. Phylogenetic analysis
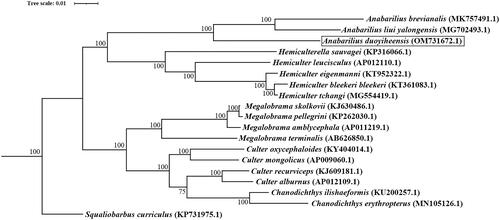
The model of GTR + I + G was the perfect substitution model for phylogenetic construction. The ML tree showed that A. duoyiheensis is phylogenetically closest to the clade of A. liui yalongensis and A. brevianalis. The genus of Anabarilius was monophyletic, and a sister group to clade of genus Hemiculter ().
Figure 3. Phylogenetic tree of A. duoyiheensis and other 18 species based on the complete mitochondrial sequences. Squaliobarbus curriculus was set to be outgroup. The bootstrap values were marker near the nodes. Accession numbers for each species were shown after the name of the species. The A. duoyiheensis generated in this study (OM731672.1) was labeled in black outline.
4. Discussion and conclusions
In this study, we first presented the mitochondrial genome of A. duoyiheensis, which is also the first molecular biological data for this species. The availability of this mitochondrial genome will be essential for future DNA barcoding, taxonomic, and phylogenetic studies of the family Xenocyprididae, and also valuable for resource conservation.
Ethical approval
The collection of specimens conformed to the Ethics Committee for Animal Experiments of Yunnan Institute of Fishery Sciences Research. The sample in this study did not involve protected and endangered animals. Obtaining the sample was legal and the process of this research was in line with the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Junjie Wu, Fangpeng Jin, Xiangxing Lu, and Yun Leng conceived this study and wrote the manuscript together. Junjie Wu, Fangpeng Jin, Hongman Yu, Jingxia Zhao, Pengxiang Zuo, Xiangxing Lu, and Jianyu Song carried out the experiments and analyzed the data. Yun Leng took the photograph of specimen. Junjie Wu, Xing Liu, Fangpeng Jin, Jingxia Zhao, Yun Leng, and Zhifei Wang contributed to the collection of specimen of A. duoyiheensis. Jianyu Song and Xiangxing Lu are responsible for the critical revision of professional knowledge content and final approval of the version to be published.
Supplemental Material
Download TIFF Image (57.1 MB)Supplemental Material
Download MS Word (12.1 KB)Acknowledgements
The authors acknowledge the Tsingke Biotechnology Co., Ltd. for bioinformatic support. We thank the editors and reviewers for their valuable comments.
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The complete mitochondrial genome assembly data were available in GenBank database under the accession number OM731672.1 (https://www.ncbi.nlm.nih.gov/nuccore/OM731672.1/). The associated BioProject, BioSample, and SRA numbers are PRJNA909186, SAMN32069822, and SRR22548207, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Chen H, Wang D, Duan X, Liu S, Chen D. 2019. The mitochondrial genome of Culter oxycephaloides (Cypriniformes, Cyprinidae). Mitochondrial DNA B Resour. 4(2):2392–2393. doi: 10.1080/23802359.2017.1383196.
- Chen L, Li B, Zhou L, Zhao G. 2016. The complete mitochondrial genome sequence of Predatory carp Chanodichthys erythropterus (Cypriniformes: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1119–1120. doi: 10.3109/19401736.2014.933328.
- Chen X. 2013. Checklist of fishes of Yunnan. Dongwuxue Yanjiu. 34(4):281–343. doi: 10.11813/j.issn.0254-5853.2013.4.0281.
- Dai X, Li W, Tian S. 2016. Complete mitochondrial genome of Hemiculter bleekeri bleekeri. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4338–4339. doi: 10.3109/19401736.2015.1089490.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 537:113–137. doi: 10.1007/978-1-59745-251-9_6.
- Guo G, Yi S, Wang W. 2017. The complete mitochondrial genome of Yihe bream, Megalobrama amblycephala Yih. Mitochondrial DNA B Resour. 2(2):566–567. doi: 10.1080/23802359.2017.1365652.
- He B, Chen Y, Liu Y, Du J, Deng X. 2016. The complete mitochondrial genome of Hemiculterella sauvagei (Teleostei, Cyprinidae, Hemiculterella). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3322–3324. doi: 10.3109/19401736.2015.1018204.
- He Y, Gong J, Zhu T, Wu X, Zhu Y, Yang D. 2019. Complete mitochondrial genome and phylogenetic analysis of Anabarilius brevianalis (Teleostei, Cyprinidae, Cultrinae). Mitochondrial DNA Part B. 4(1):1834–1836. doi: 10.1080/23802359.2019.1612717.
- Imoto JM, Saitoh K, Sasaki T, Yonezawa T, Adachi J, Kartavtsev YP, Miya M, Nishida M, Hanzawa N. 2013. Phylogeny and biogeography of highly diverged freshwater fish species (Leuciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene. 514(2):112–124. doi: 10.1016/j.gene.2012.10.019.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9(4):299–306. doi: 10.1093/bib/bbn017.
- Li S, Liu X, Bai Y, Huang T, Wang J, Qu H, Chen L, Jiang W. 2016. The complete mtDNA genome of Erythroculter ilishaeformis: genome characterization and phylogenetic analysis. Mitochondrial DNA B Resour. 1(1):39–40. doi: 10.1080/23802359.2015.1137812.
- Li W, Mao W, Lu Z. 2002. A new species of Cyprinidae from Yunnan. J Zhanjiang Ocean Univ. 22(1):1–2.
- Liu K, Ma HJ, Feng XY, Xie N. 2020. Complete mitochondrial genome of the hybrid of Culter alburnus (♀) × Megalobrama terminalis (♂). Mitochondrial DNA B Resour. 5(3):2316–2317. doi: 10.1080/23802359.2020.1772690.
- Liu X, Xiao K, Zhao X, Liu J, Zhang X, Guo W, Chen L, Du H. 2016. The complete mitochondrial genome of the Megalobrama pellegrini (Teleostei: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3069–3070. doi: 10.3109/19401736.2014.1003913.
- Luo C, Chen P, Zhu J, Ma Z. 2022. The complete mitogenome of Hemiculter leucisculus (Basilewsky, 1855) in Hainan Island and its phylogenetic status. Mitochondrial DNA B Resour. 7(2):396–398. doi: 10.1080/23802359.2022.2040390.
- Modi A, Vai S, Caramelli D, Lari M. 2021. The Illumina sequencing protocol and the NovaSeq 6000 system. Methods Mol Biol. 2242:15–42. doi: 10.1007/978-1-0716-1099-2_2.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256. doi: 10.1093/molbev/msn083.
- Rivero ER, Neves AC, Silva-Valenzuela MG, Sousa SO, Nunes FD. 2006. Simple salting-out method for DNA extraction from formalin-fixed, paraffin-embedded tissues. Pathol Res Pract. 202(7):523–529. doi: 10.1016/j.prp.2006.02.007.
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63(6):826–841. doi: 10.1007/s00239-005-0293-y.
- Yang H, Li Q, Shu H, Yang L, Wu X, Yue L, Hou L. 2016. The complete mitochondrial genome of the Megalobrama skolkovii (Cyprinidae: Cultrinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):736–737. doi: 10.3109/19401736.2014.913165.
- Yang H, Zhao H, Sun J, Xie Z, Yang Z, Liu L. 2016. The complete mitochondrial genome of the Culter recurviceps (Teleostei, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):762–763. doi: 10.3109/19401736.2014.915529.
- Yang J, Chu X. 1987. Phylogeny of the Cyprinid genus Anabarilius (Pisces: Cyprinidae). Zool Res. 8(3):261–276.
- Zeng L, Yang K, Wen A, Xie M, Yao Y, Xu H, Zhu G, Wang Q, Jiang Y, He T, et al. 2019. Complete mitochondrial genome of the endangered fish Anabarilius liui yalongensis (Teleostei, Cyprinidae, Cultrinae). Mitochondrial DNA B Resour. 4(1):21–22. doi: 10.1080/23802359.2018.1535850.
- Zeng S, Zhou L, Wang P, Tang Q, Zeng L, Xu P, Li G. 2016. The mitochondrial genome of Squaliobarbus curriculus of Pearl River. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4217–4218. doi: 10.3109/19401736.2015.1022747.
- Zou Y, Liu T, Li Q, Wen Z, Qin C, Li R, Wang D. 2019. Complete mitochondrial genome of Hemiculter tchangi (Cypriniformes, Cyprinidae). Conserv Genet Resour. 11(1):1–4. doi: 10.1007/s12686-017-0949-0.