Abstract
Tethea albicostata is a widely distributed insect species in northern and central China. To date, few studies have been conducted on this species, with the exception of morphological taxonomy studies. Here, we report the complete mitochondrial genome of T. albicostata collected in China. The circular-mapping mitogenome is 15,308 bp in length, with an overall A + T content of 80.52%, encoding 2 ribosomal RNA genes, 22 transfer RNA genes, and 13 protein-coding genes. The gene arrangement and components of T. albicostata are identical to those of most other Lepidopteran insects. Phylogenetic analysis based on mitogenomes showed that T. albicostata is grouped with Drepana pallida, which belongs to the same family as Drepanidae. The family Drepanidae formed a separate branch from other families in the phylogenetic tree. This study determined the second mitochondrial genome of the Drepanidae species.
Introduction
Tethea albicostata (Bremer 1861) is a member of the Thyatirinae subfamily of the family Drepanidae, which belongs to the order Lepidoptera. It is widely distributed in most areas of China, Russia, South Korea, and Japan (Kim et al. Citation2006). The larvae of Drepanidae are leaf feeders and host plants, including Anacardiaceae, Betulaceae, and Caprifoliaceae, are diverse (Heppner Citation2008). Very few studies have been conducted on this species, therefore, the knowledge base is limited to its morphological characteristics and taxonomic status (Kim et al. Citation2006; Jiang et al. Citation2015; Leley Citation2016). Currently, there is a lack of reliable identification features at the morphological level; therefore, the current classification of Drepanidae based on morphological characteristics and molecular markers is inconsistent (Rego Citation1995; Timmermans et al. Citation2014; de Chambrier et al. Citation2015; Mitter et al. Citation2017). Molecular data can provide supporting evidence for a more accurate morphological taxonomic status (Skeríková et al. Citation2001). However, the NCBI has only recorded the COI sequence of T. albicostata, and only one other mitochondrial genome (Drepana pallida) has been published under the family Drepanidae to date. In this study, we determined the complete mitochondrial genome of T. albicostata (), which represents a new genus (Tethea) of Drepanidae. Additionally, we examined the phylogenetic relationships of Drepanidae based on mitochondrial genomes.
Figure 1. Dorsal view of a male Tethea albicostata. The image is referenced from the bold systems (Ratnasingham and Hebert Citation2007). The sample ID is SDNU-INS-01482. This figure is attribution noncommercial sharing. http://v3.boldsystems.org/index.php/taxbrowser_taxonpage?taxon=Tethea+albicostata&searchTax=.

Materials
The T. albicostata sample used in this study was a moth collected in 2020 from Changchun (43°48′38″N, 125°24′14″E) in the Jilin Province of China. Identification and characterization of T. albicostata were conducted based on morphological and molecular methods. Morphological characteristics were obtained from the previous studies (Surlykke et al. Citation2003), and molecular identification was carried out using the mitochondrial COI marker (Folmer et al. Citation1994). A nucleotide BLAST search in GenBank showed that the COI sequence of the specimen had 99.67–100.00% sequence similarity with the COI sequence of T. albicostata. The COI sequences downloaded from NCBI were used to construct a neighbor-joining (NJ) phylogenetic tree for molecular identification of the sample (Supporting Information Figure S1). Voucher specimen and DNA were deposited at the Agricultural Genomics Institute in Shenzhen, China (https://www.agis.org.cn/, contact Lei Zhang, [email protected]) under voucher number AGIS-2020J22.
Methods
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Cat. no. 69506; Qiagen, Germany). DNA quality and concentration were determined using a NanoDrop One UV–Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A total of 0.5 μg of genomic DNA was used to construct a 350 bp insert library, which was then sequenced in a 2 × 150 bp format on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Mosdepth software was further used to evaluate the sequencing depth of mitochondrial genomes (Pedersen and Quinlan Citation2018).
NOVOPlasty v2.5.6 software (Dierckxsens et al. Citation2017) was used to assemble the complete mitochondrial genome based on Illumina reads. Annotation of mitochondrial genes was performed using the MITOS2 webserver (http://mitos2.bioinf.uni-leipzig.de/index.py) (Donath et al. Citation2019), and the mitochondrial genome map of T. albicostata was produced using OGDRAW v1.3.1 software (Greiner et al. Citation2019). The mitogenome sequences were aligned using MAFFT v7.455 (Katoh and Standley Citation2013). The mitochondrial genome annotation was checked via the CPGView (Liu et al. Citation2023). A phylogenetic tree was constructed based on the mitochondrial genome, including the control region, and the maximum-likelihood method with the best-fit model (GTR + F + I + G4) estimated using IQ-TREE v1.6.10 (Nguyen et al. Citation2015). Finally, 1000 bootstrap replicates were used, and the tree was visualized using iTol v6.5.2 (https://itol.embl.de/).
Results and discussion
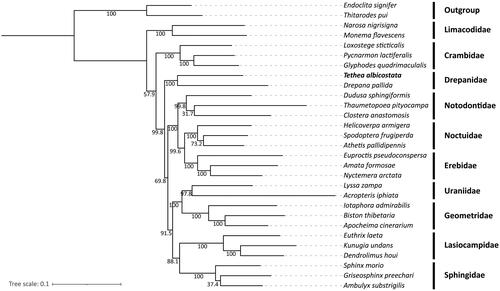
Molecular identification based on the NJ tree grouped the specimen with T. albicostata samples (Supporting Information Figure S1). The determined specimen was T. albicostata. The assembly of the mitochondrial genome was based on 142,304,470 reads (the percentage of reads with ≥Q30 score was 96.42%). The mitogenome consensus sequence coverage depth is in the range of 5112 to 32,047 (Supporting Information Figure S2), resulting in a circular molecule of 15,308 bp in length (GenBank accession no. OK149234), with a total A + T content of 80.52%, similar to that of Erebidae (80.2–81.9%), Limacodidae (80.5–81.2%), and Crambidae (79.6–81.5%). The mitogenome contains a standard set of 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNAs, and 2 ribosomal RNAs (). Gene structure analysis was performed for annotated genes, and no cis-splicing or trans-splicing genes were detected in the mitochondrial genome (Supporting Information Figure S3). The gene arrangement in the mitogenome of T. albicostata is identical to that of most Lepidopteran species (Wan et al. Citation2013). The nucleotide composition of the T. albicostata mitogenome is as follows: T, 40.27%; A, 40.25%; C, 11.61%; G, 7.87%. The negligible AT skew (0.0002) and negative GC skew (–0.1920) in the mitogenome of T. albicostata were similar to those of other published Lepidopteran species (Cameron and Whiting Citation2008). All 13 PCGs start with the ATN codons (where N represents A, T, or G), except the cox1 gene, which was initiated using the CGA sequence. This generally occurs in Lepidoptera (Cao et al. Citation2012; Chen et al. Citation2022). Most PCGs are terminated with a complete stop codon (TAA or TAG), except for cox1, cox2, and nad4. They have an incomplete stop codon (T) that can form the functional stop codon TAA via the polyadenylation processes that add 3′A residues (Ojala et al. Citation1981). In contrast to T. albicostata, five mitochondrial PCGs (cox1, cox2, nad2, nad4, and nad5) in D. pallida use an incomplete stop codon (T). The phylogenetic tree demonstrated that T. albicostata is clustered in the same branch as D. pallida, which belongs to the family Drepanidae, with 100 bootstrap values (). Among the 29 species, those from the same family clustered together, and the family Drepanidae formed a separate branch form other family in the phylogenetic tree.
Figure 2. The complete mitochondrial genome map of Tethea albicostata. Genes belonging to different functional groups are color coded. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. The inner circle represents the GC% along the mitochondrion.
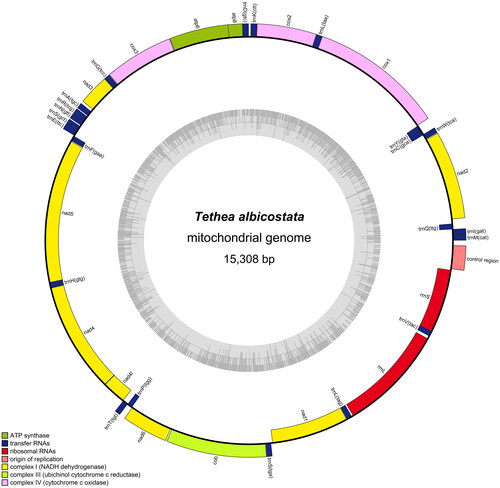
Figure 3. Maximum-likelihood (ML) tree of 29 Lepidopteran species based on the complete mitogenome sequences. Endoclita signifer and Thitarodes pui were used as outgroups. The numbers below the branches were the bootstrap values. A summary of the 29 Lepidoptera species used in the phylogenetic tree is shown in Supporting Information Table S1.
Conclusion
In this study, we present the complete mitochondrial genome of T. albicostata, the second species of Drepanidae. However, current molecular data for Drepanidae are very limited, especially mitochondrial genome-wide data, which may help resolve the taxonomic inconsistency between the molecular and morphological identification of species in this family more accurately. In summary, the mitogenome complements the current molecular data for T. albicostata and may contribute to further phylogenetic analyses of Drepanidae species.
Ethical approval
Tethea albicostata is a common insect distributed in China. Therefore, no ethical approval or other relevant permission can be provided for the study.
Authors’ contributions
Xinyue Liang and Lei Zhang collected and identified the specimens; Xinyue Liang and Ping Wang analyzed the data; Xinyue Liang wrote the manuscript; Lei Zhang and Zaiyuan Li revised the manuscript. Yutao Xiao conceived and designed the project, and finally, approved the version to be published. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (6.5 MB)Supplemental Material
Download MS Word (222.9 KB)Supplemental Material
Download MS Word (3.6 MB)Supplemental Material
Download MS Word (22.2 KB)Supplemental Material
Download PDF (310.2 KB)Disclosure statement
No potential conflict of interest is reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OK149234. The associated BioProject, BioSample, and SRA numbers are PRJNA810316, SAMN26244873, and SRR18148899, respectively.
Additional information
Funding
References
- Cameron SL, Whiting MF. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 408(1–2):112–123. doi: 10.1016/j.gene.2007.10.023.
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genom. 13(1):276.
- Chen Q, Chen L, Liao CQ, Wang X, Wang M, Huang GH. 2022. Comparative mitochondrial genome analysis and phylogenetic relationship among Lepidopteran species. Gene. 830:146516. doi: 10.1016/j.gene.2022.146516.
- de Chambrier A, Waeschenbach A, Fisseha M, Scholz T, Mariaux J. 2015. A large 28S rDNA-based phylogeny confirms the limitations of established morphological characters for classification of proteocephalidean tapeworms (Platyhelminthes, Cestoda). Zookeys. 500(500):25–59. doi: 10.3897/zookeys.500.9360.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3(5):294–299.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Heppner JB. 2008. Hooktip moths (Lepidoptera: Drepanidae). In: Capinera JL, editors. Encyclopedia of entomology. Dordrecht: Springer.
- Jiang N, Yang C, Xue D, Han H. 2015. An updated checklist of Thyatirinae (Lepidoptera, Drepanidae) from China, with descriptions of one new species. Zootaxa. 3941(1):1–48. doi: 10.11646/zootaxa.3941.1.1.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim MY, Lee HK, Ronkay L, Park KT. 2006. A review of the Korean Thyatiridae (Lepidoptera), including the Mt. Changbai-Shan. J Asia-Pac Entomol. 9(3):203–221. doi: 10.1016/S1226-8615(08)60293-9.
- Leley. 2016. Annotated catalogue of the insects of Russian Far East. Volume II. Lepidoptera Cat Ins Russian Far East. 2:1–812.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 1–11.
- Mitter C, Davis DR, Cummings MP. 2017. Phylogeny and evolution of Lepidoptera. Annu Rev Entomol. 62(1):265–283. doi: 10.1146/annurev-ento-031616-035125.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474. doi: 10.1038/290470a0.
- Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 34(5):867–868. doi: 10.1093/bioinformatics/btx699.
- Ratnasingham S, Hebert PD. 2007. Bold: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x.
- Rego AA. 1995. A new classification to the cestode order Proteocephalidea Mola. Rev Bras Zool. 12(4):791–814. doi: 10.1590/S0101-81751995000400009.
- Skeríková A, Hypsa V, Scholz TV. 2001. Phylogenetic analysis of European species of Proteocephalus (Cestoda: Proteocephalidea): compatibility of molecular and morphological data, and parasite-host coevolution. Int J Parasitol. 31(10):1121–1128. doi: 10.1016/s0020-7519(01)00226-0.
- Surlykke A, Yack JE, Spence AJ, Hasenfuss I. 2003. Hearing in hooktip moths (Drepanidae: Lepidoptera). J Exp Biol. 206(Pt 15):2653–2663. doi: 10.1242/jeb.00469.
- Timmermans MJ, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178. doi: 10.1016/j.ympev.2014.05.031.
- Wan X, Kim MJ, Kim I. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40(11):6333–6349. doi: 10.1007/s11033-013-2748-3.
