Abstract
Micranthes octopetala (Nakai) Y.I.Kim & Y.D. Kim et al. 2015, which belongs to the family Saxifragaceae, is a perennial herb endemic to Korea. M. octopetala was originally treated as a synonym of M. manchuriensis. However, in 2015, molecular phylogenetic analysis confirmed that M. octopetala is an independent species. In this study, the plastid genome of M. octopetala was sequenced for the first time, and the taxonomic position of this species was identified. The complete plastid genome of M. octopetala has a total length of 149 751 bp (large single copy: 83 083 bp; small single copy: 17 196 bp; inverted repeat: 24 736 bp), containing 130 genes, including 79 CDS, 30 tRNAs, and 4 rRNAs. Moreover, the absence of intron in the rpl2 gene, which is a common feature of Saxifragaceae, was confirmed. Phylogenetic analysis based on 79 protein-coding genes from 21 species revealed that M. octopetala belongs to the genus Micranthes, being a sister to other Micranthes species. The plastid genome of M. octopetala obtained in this study provides fundamental information for future studies on the genus Micranthes.
Introduction
Micranthes Haw., which belongs to the family Saxifragaceae, is an herbaceous plant genus mainly distributed in the Northern Hemisphere, consisting of approximately 80 species. It previously belonged to the genus Saxifraga L. but has been recognized as an independent genus through several molecular phylogenetic studies based on chloroplast DNA genes or internal transcribed spacer (ITS) regions in its genome (Soltis et al. Citation1996; Citation2001; Xiang et al. Citation2012; Deng et al. Citation2015). Understanding the taxonomic position of Micranthes within Saxifragaceae can aid in comparative studies with related genera and contribute to our understanding of the evolutionary history within the family (Deng et al. Citation2015).
Micranthes octopetala (Nakai) Y.I.Kim & Y.D. Kim et al. Citation2015, which is endemic to Korea, was treated as an independent species in 2015 by Kim et al. (Citation2015). M. octopetala is a perennial plant that typically attains a height of 15–30 cm and grows on the surface of rocks in the shade of deep mountainous areas. Despite controversies regarding the taxonomic position of M. octopetala, this has been majorly resolved in a previous study (Kim et al. Citation2015). M. octopetala was originally included in the genus Saxifraga (Saxifraga octopetala Nakai 1918), but Kim et al. (Citation2015) revealed that M. octopetala belongs to the genus Micranthes and proposed a new combination-specific name, Micranthes octopetala. Although M. octopetala was ambiguously treated as M. manchuriensis, according to Kim et al. (Citation2015), a phylogenetic tree based on the nrITS region demonstrated that M. octopetala and M. manchuriensis formed an independent clade. In particular, M. octopetala was shown to be phylogenetically close to M. nelsoniana and M. manchuriensis, with M. manchuriensis forming a sister group with M. nelsoniana var. pacifica and M. fusca. In addition, M. octopetala is distinguished from M. manchuriensis by its distinct morphological features, such as the presence of an underground stolon.
Plastid genomes are often used for phylogenetic analysis because of their high conservation and slow evolutionary rate compared with other genetic materials (Provan et al. Citation2001; Ravi et al. Citation2008). However, to date, the plastid genome of M. octopetala has remained unknown. To the best of our knowledge, the plastid genome of M. octopetala was assembled and presented for the first time in this study, and phylogenetic relationships within the family Saxifragaceae were identified.
Materials and methods
Leaves of M. octopetala were collected at Mt. Gwangdeoksan, Hwacheon-gun, Gangwon-do province, South Korea (38°06′55.8″N 127°25′52.3″E) () and voucher specimens were deposited in the Korea National Arboretum (http://www.nature.go.kr/kbi/plant/smpl/KBI_2001_030100.do, Hee-Young Gil, E-mail: [email protected]) under the voucher number (Hyh144). Total genomic DNA was extracted from dried leaves using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) with silica gel. The extracted gDNA was subjected to next-generation sequencing (NGS) using a MiSeq sequencing system (Illumina, Seoul, Korea), and a total of 9 992 786 reads were obtained. To remove poor-quality reads, the raw data were imported and trimmed using Geneious Prime (Kearse et al. Citation2012) with a 5% error probability limitation. Using M. melanocentra (GenBank accession = NC_056191.1) as a reference, the "map to reference" option and De novo assembly were used so that only raw reads related to the plastid genome were selected and assembled according to the reference (Kearse et al. Citation2012). The read coverage depth of the assembled genome was 347.21× (Supplementary Figure 1). The gene content and order of the plastome of M. octopetala were annotated using GeSeq and scrutinized using Geneious Prime and tRNA scans (Kearse et al. Citation2012; Tillich et al. Citation2017; Chan and Lowe Citation2019). Finally, the OGDraw program was used to visualize the plastid genome of M. octopetala (Greiner et al. Citation2019).
Figure 1. Micranthes octopetala in its Native environment at Mt. Gwangdeoksan, Hwacheon-gun, Gangwon-do province, South Korea. The species reference image was taken by Sa-Bum Jang. M. octopetala is a perennial herb that grows to 15–30 cm and has 1–4 leaves. Flowers with 8 white petals bloom from June to July. Flowering stems are erect and leafless. The characteristic that can be distinguished from similar species (M. manchuriensis) is that it has an underground stolon.

For phylogenetic analysis, 18 species of Saxifragaceae were used, with 3 closely related species of Saxifragaceae (Ribes glaciale, GenBank accession number MK887908; Itea chinensis, GenBank accession number MH191391; Penthorum chinense, GenBank accession number JX436155) being used as outgroups. Using MAFFT, 79 protein-coding genes were extracted from the plastid genome, aligned, and combined to generate molecular data (Katoh and Standley Citation2013; Zhang et al. Citation2020). Maximum likelihood (ML) analysis was performed based on the concatenated molecular data using the IQ-tree software of the PhyloSuite program (Nguyen et al. Citation2015; Zhang et al. Citation2020). According to ModelFinder in PhyloSuite, the best-fit model was GTR + F + I + G4, and an ML tree with 5000 bootstrap replications was constructed (Zhang et al. Citation2020).
Results
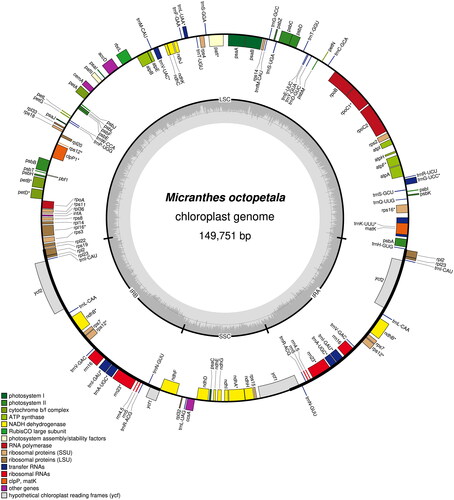
We confirmed that the total length of the plastid genome of M. octopetala was 149 751 bp, with 37.8% GC content (). It exhibited the typical quadripartite structure of angiosperms, with the lengths of the large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR) regions being 83 083, 17 196, and 24 736 bp, respectively. We also found that the GC content of the IR region (43.3%) was higher than that of the LSC (35.7%) and SSC regions (31.7%). We identified a total of 130 genes in M. octopetala plastid genes, including repetitive genes in the IR region, consisting of 79 CDS, 30 tRNAs, and 4 rRNAs (Supplementary Table 1). We detected that 14 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained 1 intron, whereas 2 coding genes (clpP1 and pafI) had 2 introns (Supplementary Figure 2). In addition, we found that rps12 is a trans-splicing gene located in the IRs and LSC regions (Supplementary Figure 3). We further confirmed that the intron of rpl2 was deleted.
Figure 2. The complete plastid genome of Micranthes octopetala. Graphic showing features of its plastome was generated using OGDraw. Colored boxes represent functional groups of plastid-coding genes. The clockwise and counter-clockwise transcribed genes are drawn inside and outside of the circle, respectively. The small gray bar graphs in the inner circle show the GC contents.
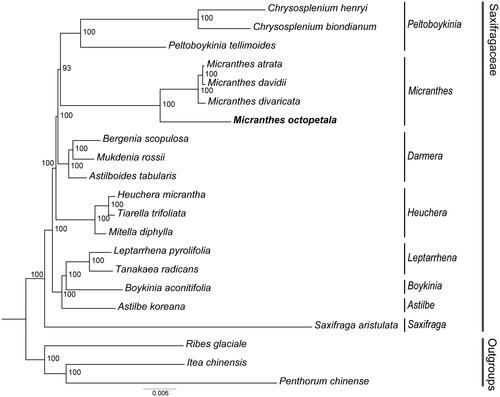
The ML tree with GTR + F + I + G4 model constructed based on the 79 concatenated protein-coding genes showed that Saxifragaceae is a monophyletic family, with most nodes exhibiting a high support value (), as described in previous studies (Soltis et al. Citation2001; Deng et al. Citation2015). We determined that M. octopetala belongs to the genus Micranthes and was positioned as a sister to other Micranthes species. We confirmed that the group closest to the Micranthes group was the Peltoboykinia group (Chrysosplenium, Peltoboykinia).
Figure 3. The ML tree of 18 species of Saxifragaceae based on concatenated protein-coding genes. M. octopetala is marked in bold. Bootstrap support values shown at the branches. The following sequences were used: Chrysosplenium henryi OK336532, Chrysosplenium biondianum OK336542, Peltoboykinia tellimoides MZ779205 (Yang et al. Citation2022), Micranthes atrata ON720899 (Yuan et al. Citation2023), Micranthes davidii ON720901 (Yuan et al. Citation2023), Micranthes divaricata ON720902 (Yuan et al. Citation2023), bergenia scopulosa KY412195 (Bai et al. Citation2018), mukdenia rossii MG470844 (Liu et al. Citation2018), astilboides tabularis MT316511 (Ha et al. Citation2020), heuchera micrantha OL489769, tiarella trifoliata MH708572, mitella diphylla MH708564, leptarrhena pyrolifolia MN496070 (Folk et al. Citation2020), tanakaea radicans MW300581 (Su et al. Citation2021), boykinia aconitifolia MN496058 (Folk et al. Citation2020), astilbe koreana MK990830, Saxifraga aristulata ON720851 (Yuan et al. Citation2023), Ribes glaciale MK887908, Itea chinensis MH191391 (Dong et al. Citation2018), and Penthorum chinense JX436155 (Dong et al. Citation2013).
Discussion and conclusions
In this study, the plastome of M. octopetala, a Korean endemic species, was assembled and its phylogenetic relationships were confirmed. The plastid genome length of M. octopetala was determined to be 149 751 bp, containing a total of 130 genes. The absence of introns in the rpl2 gene of M. octopetala, which is a characteristic commonly observed in Saxifragaceae plants, was demonstrated (Downie et al. Citation1991; Dong et al. Citation2013; Liu et al. Citation2020). Gene losses occur frequently during the process of plant evolution and provide evidence for elucidating the evolutionary history of plants (Jansen et al. Citation2007; Han et al. Citation2022). In the case of M. octopetala, the loss of the rpl2 intron can be attributed to homologous recombination and gene conversion mechanisms, which represent the most plausible causal factors (Gu et al. Citation2016; Ding et al. Citation2019; Han et al. Citation2022). Phylogenetic analysis based on the plastid genome was performed to infer phylogenetic relationships between M. octopetala and related taxa. Similar to the results of a previous study, M. octopetala was identified to belong to the genus Micranthes and was thus located as a sister group to other Micranthes species (Kim et al. Citation2015). In this study, we obtained the plastid genome of M. octopetala, which will aid future phylogenetic and evolutionary studies of the genus Micranthes.
Ethical approval
The materials used in this study did not involve ethical conflicts. Therefore, no specific permission or license was required.
Authors’ contributions
Tae-Hee analyzed the data and drafted the manuscript. Young-Ho collected plant materials and performed the experiments. Sang-Chul analyzed the data and revised the manuscript. Hyuk-Jin conceived of and designed the original study. All authors agreed to the submitted version of the manuscript.
Supplemental Material
Download JPEG Image (669.8 KB)Supplemental Material
Download JPEG Image (698.4 KB)Supplemental Material
Download JPEG Image (894.6 KB)Supplemental Material
Download MS Excel (12.3 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study are available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ835312. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA956675, SRR24198730, and SAMN34227787, respectively.
Additional information
Funding
References
- Bai G, Fang L, Li S, Cui X. 2018. Characterization of the complete chloroplast genome sequence of Bergenia scopulosa (Saxifragales: Saxifragaceae). Conservation Genet Resour. 10(3):363–366. doi: 10.1007/s12686-017-0825-y.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Deng J-b, Drew BT, Mavrodiev EV, Gitzendanner MA, Soltis PS, Soltis DE. 2015. Phylogeny, divergence times, and historical biogeography of the angiosperm family Saxifragaceae. Mol Phylogenet Evol. 83:86–98. doi: 10.1016/j.ympev.2014.11.011.
- Ding H, Zhu R, Dong J, Bi D, Jiang L, Zeng J, Huang Q, Liu H, Xu W, Wu L, et al. 2019. Next-generation genome sequencing of Sedum plumbizincicola sheds light on the structural evolution of plastid rRNA operon and phylogenetic implications within saxifragales. Plants. 8(10):386. doi: 10.3390/plants8100386.
- Dong W, Xu C, Cheng T, Lin K, Zhou S. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5(5):989–997. doi: 10.1093/gbe/evt063.
- Dong W, Xu C, Cheng T, Zhou S. 2013. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLOS One. 8(10):e77965. doi: 10.1371/journal.pone.0077965.
- Dong W, Xu C, Wu P, Cheng T, Yu J, Zhou S, Hong DY. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330. doi: 10.1016/j.ympev.2018.04.033.
- Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, Watson JC, Palmer JD. 1991. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution. 45(5):1245–1259. doi: 10.2307/2409731.
- Folk RA, Sewnath N, Xiang CL, Sinn BT, Guralnick RP. 2020. Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae). BMC Plant Biol. 20(1):324. doi: 10.1186/s12870-020-02533-x.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Gu C, Tembrock LR, Johnson NG, Simmons MP, Wu Z. 2016. The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae). PLOS One. 11(3):e0150752. doi: 10.1371/journal.pone.0150752.
- Ha YH, Gil HY, Kim DK. 2020. First report of complete chloroplast genome sequence of monotypic genus Astilboides (Saxifragaceae) in North East Asia. Mitochondrial DNA Part B Resour. 5(3):3598–3599. doi: 10.1080/23802359.2020.1824595.
- Han S, Ding H, Bi D, Zhang S, Yi R, Gao J, Yang J, Ye Y, Wu L, Kan X. 2022. Structural diversities and phylogenetic signals in plastomes of the early-divergent angiosperms: a case study in Saxifragales. Plants. 11(24):3544. doi: 10.3390/plants11243544.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104(49):19369–19374. doi: 10.1073/pnas.0709121104.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kim Y-I, Cho SH, Kim B-Y, Lee JH, Kang D-H, Kim S, Doudkin RV, Kim Y-D. 2015. A new combination for Saxifraga octopetala (Saxifragaceae) and its phylogenetic relationship. Korean J Pl Taxon. 45(4):306–317. doi: 10.11110/kjpt.2015.45.4.306.
- Liu L-X, Du Y-X, Folk RA, Wang S-Y, Soltis DE, Shang F-D, Li P. 2020. Plastome evolution in Saxifragaceae and multiple plastid capture events involving Heuchera and Tiarella. Front Plant Sci. 11:361. doi: 10.3389/fpls.2020.00361.
- Liu L, Wang Y, He P, Li P, Lee J, Soltis DE, Fu C. 2018. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics. 19(1):1–17. doi: 10.1186/s12864-018-4633-x.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Provan J, Powell W, Hollingsworth PM. 2001. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol. 16(3):142–147. doi: 10.1016/s0169-5347(00)02097-8.
- Ravi V, Khurana J, Tyagi A, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271(1-2):101–122. doi: 10.1007/s00606-007-0608-0.
- Soltis DE, Kuzoff RK, Conti E, Gornall R, Ferguson K. 1996. matK and rbcL gene sequence data indicate that Saxifraga (Saxifragaceae) is polyphyletic. Am J Bot. 83(3):371–382. doi: 10.1002/j.1537-2197.1996.tb12717.x.
- Soltis DE, Kuzoff RK, Mort ME, Zanis M, Fishbein M, Hufford L, Koontz J, Arroyo MK. 2001. Elucidating deep-level phylogenetic relationships in Saxifragaceae using sequences for six chloroplastic and nuclear DNA regions. Ann Mo Bot Gard. 88(4):669–693. doi: 10.2307/3298639.
- Su T, Luo GL, Gu L, Liao HM, Hu GX. 2021. The complete plastome sequence of Tanakaea radicans (Saxifragaceae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(3):946–947. doi: 10.1080/23802359.2021.1889409.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Xiang C-L, Gitzendanner MA, Soltis DE, Peng H, Lei L-G. 2012. Phylogenetic placement of the enigmatic and critically endangered genus Saniculiphyllum (Saxifragaceae) inferred from combined analysis of plastid and nuclear DNA sequences. Mol Phylogenet Evol. 64(2):357–367. doi: 10.1016/j.ympev.2012.04.010.
- Yang R, Li R, Dong M, Liu L. 2022. Characterization of the complete chloroplast genome of Peltoboykinia tellimoides (Saxifragaceae). Mitochondrial DNA Part B Resour. 7(5):741–743. doi: 10.1080/23802359.2022.2070040.
- Yuan R, Ma X, Zhang Z, Gornall RJ, Wang Y, Chen S, Gao Q. 2023. Chloroplast phylogenomics and the taxonomy of Saxifraga section Ciliatae (Saxifragaceae). Ecol Evol. 13(1):e9694. doi: 10.1002/ece3.9694.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
