Abstract
The Hainan Island Shrew Crocidura wuchihensis is a small-bodied insectivore species belonging to the Soricidae family. In this study, we determined its the complete mitochondrial genome. The whole mitochondrial genome was found to be 17,253 bp in length and comprises 13 protein-coding genes, 22 tRNAs, 2 rRNAs and a control region. The base composition of the C. wuchihensis total mitogenome as follows: A, 32.8%; G, 13.0%; T, 32.1%; and C, 22.1%, with an A + T content of 64.9%. Notably, a tandem repeat sequence (5′-CAC ACG TGT ACA-3′) was identified in the control region with 24 copy numbers. Additionally, phylogenetic analysis indicated that C. wuchihensis is closely related to Crocidura tanakae and Crocidura dongyangjiangensis based on the concatenated sequences of the 13 protein-coding genes. The characterization of the shrew’s mitogenome will provide the foundation for its use in population genomics and systematic studies of Soricidae.
1. Introduction
The Hainan Island Shrew Crocidura wuchihensis Shaw et al. (Citation1966) is a small-bodied insectivore belonging to the family Soricidae. It inhabits regions across Hainan, Yunnan, and Guangxi provinces in China as well as in Vietnam (Smith et al. Citation2009; Chen et al. Citation2020; Wei et al. Citation2022). The genus Crocidura exhibits a remarkably conserved external morphology, often leading to taxonomic confusion (Chen et al. Citation2020). Although more than 170 species fall under this genus (Wilson and Reeder Citation2005); the complete mitochondrial genome sequences have been reported for < 20 species among them. To the best of our knowledge, sequences of only partial fragments of mitochondrial DNA (Cytb, ND2, and Cox1) are available for C. wuchihensis. Given this gap, we aimed to sequenced the complete mitochondrial genome of C. wuchihensis to understand phylogenetic relationships within Soricidae family.
2. Materials and methods
2.1. Species collection
The deceased animal () was collected from the Jian Feng Mountains (109.289°E, 18.878°N), Hainan, China, and subsequently preserved in the animal specimen room of Hainan Normal University (Feiyun Tu, [email protected]) with the voucher number JFL-015.
2.2. Methods
A muscle sample was extracted from the preserved specimen, and the total genomic DNA was isolated from the muscle tissue using a standard phenol-chloroform extraction protocol (Sambrook et al. Citation1989). The genome was sequenced using Sanger sequencing using the primer walking approach (Table S1). The complete mitochondrial genome sequence was then assembled and manually refined utilizing SeqMan (DNASTAR 7.1.0) (Swindell and Plasterer Citation1997) following analysis of chromatogram files. The strategy view of the assembled sequence (Figure S1) was then generated with SeqMan (DNASTAR 7.1.0). Subsequently, the mitogenomic sequences were imported into the MITOS web servers for the preliminary determination of gene boundaries. To identify precise positions of protein-coding genes (PCGs), open reading frames were identified(ORFs) using the vertebrate mitochondrial genetic code. Identification of all tRNAs was performed using MITOS2 (Donath et al. Citation2019). Accurate delineations of tRNAs, 12S rRNA, and 16S rRNA were established through comparison with mitochondrial sequences of related species (NC_046831 and NC_026204). The circular map of the mitochondrial genome was constructed using SeqBuilder software.
To detect repetitive tandem sequences, the control region sequences of 12 Crocidura species were examined using the web server Tandem Repeat Finder (https://tandem.bu.edu/trf/advanced_submit) with the default settings, except the parameter “Minimum alignment score to report repeat” which was set to the maximum value of 150 (Benson Citation1999). The identified tandem repeat sequences and their copy number were detected and given in Table S2. The complete mitogenome sequences of 19 Crocidura species (Table S3) were retrieved from GenBank. By amalgamating these sequences with the mitogenome sequenced in this study, we generated concatenated datasets involving the 13 PCGs. To depict the evolutionary relationships, a phylogenetic tree was constructed using MEGA6.0 (Tamura et al. Citation2013) and based on the concatenated datasets, utilizing the Kimura 2-parameter model.
3. Results
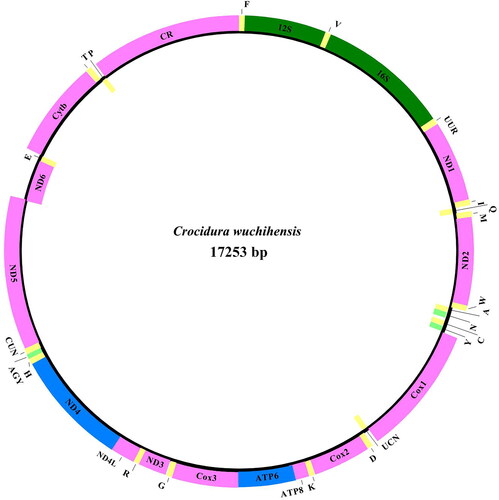
The whole mitochondrial genome of C. wuchihensis is 17,253 base pairs (bp) in length and contains 13 PCGs, 22 tRNAs, 2 rRNAs, and a control region (). The base composition of the C. wuchihensis mitogenome was found to be: A, 32.8%; G, 13.0%; T 32.1%; and C 22.1%. Moreover, a notable A + T-rich pattern (64.9%) was observed. Eleven PCGs begin with the standard ATG, while ND2 and ND3 initiate with ATT and ATA, respectively. PCGs contained both complete termination codons (TAA for ND1, Cox1, Cox2, ATP8, ATP6, ND4L, ND5, ND6; TAG for ND2, ND3; AGA for Cytb) and an incomplete termination codon (T for Cox3, ND4).
Figure 2. Gene map of the mitochondrial genome of C. wuchihensis. Most mitochondrial genes (encoding on the H-strand) were located outside of the circle. Eight tRNA genes (encoding on the L-strand) were located within the circle. The color of 12 mitochondrial genes were labeled as pink. ATP6 and ND4 were labeled as blue. Two rRNAs (12S and 16S) were labeled as green. Three tRNAs (N, Y, and AGY) were labeled as light green, while the remaining tRNAs were labeled as yellow. F: tRNA-Phe, V: tRNA-Val, UUR: tRNA-Leu1, I: tRNA-Ile, Q: tRNA-Glu; M: tRNA-Met, W: tRNA-Trp; A: tRNA-Ala; N: tRNA-Asp; C: tRNA-Cys; Y: tRNA-Tyr; UCN: tRNA-Ser1; D: tRNA-Asn; K: tRNA-Lys; G: tRNA-Gly; R: tRNA-Arg; H: tRNA-His; AGY: tRNA-Ser2; CUN: tRNA-Leu2; E: tRNA-Gln; T: tRNA-Thr; P: tRNA-Pro.
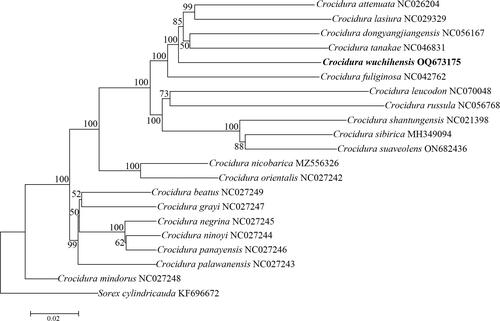
The 12S rRNA and 16S rRNA genes of C. wuchihensis are 969 bp and 1,571 bp in length, respectively. The 12S rRNA gene is situated between the tRNA-Phe and tRNA-Val genes, with a nucleotide composition of A 36.1%, T 26.3%, C 20.4%, and G 17.2%, while the 16S rRNA gene is located between tRNA-Val and tRNA-Leu, with exhibits a nucleotide content of A 37.6%, T 26.9%, C 18.9%, and G 16.6%. The two rRNA genes are AT-rich (12S rRNA = 62.4% and 16S rRNA = 64.5%). The control region gene is 1,799 bp in length. In the control region of all 12 Crocidura species, tandem repetitions were found (Table S2). Notably, a tandem repeat sequence [5′-(CAC ACG TGT ACA)24-3′ motif] was found in the C. wuchihensis regulatory region sequence, starting at position 1,277 and ending at position 1,566. Additionally, four copies of the consensus pattern (79 bp) were found in the control region; however, the sequences of these four copies were not completely consistent (Table S2). Furthermore, the identical tandem repeats (5′-(CAC ACG TGT ACA)30-3′) were also found in Crocidura russula (NC056768) (Table S2). According to the phylogenetic tree, C. wuchihensis belonged to the larger clade of Crocidura species and share the most recent common ancestor with four congeneric species: C. tanakae, C. dongyangjiangensis, C. attenuata, and C. lasiura (). C. tanakae and C. dongyangjiangensis share a recent common ancestor with a low support value.
4. Discussion and conclusion
The C. wuchihensis mitochondrial genome contains 13 PCGs, 22 tRNAs, 2 rRNAs, and a control region (). The composition is similar to the mitochondrial genome of related species (Li et al. Citation2021). According to the phylogenetic analysis, C. wuchihensis was shown to be closely related to C. tanakae and C. dongyangjiangensis forming a sibling relationship, which was consistent with the results of previous studies (Chen et al. Citation2021; Li et al. Citation2021). Previously, Chen et al. (Citation2020) reported a close relationship between C. wuchihensis and C. tanakae based on the multilocus data (mtDNA + nDNA, approximately 3,300 bp). Similarly, Liu et al. (Citation2020) proposed a sister relationship between C. wuchihensis and C. indochinesis using single Cytb data. However, the results of our study diverge from the findings of Liu et al. (Citation2020). This divergence can be attributed to differences in taxon sampling.
In the present study, we identified identical tandem repeats that were detected within both C. wuchihensis (OQ673175) and C. russula (NC056768). However, these repeats were notably absent in the control region sequences of closely related species C. tanakae and C. dongyangjiangensis (Table S2). This observation implies that the presence or absence of these tandem repeats may not accurately reflect the species phylogeny (Song et al. Citation2015; Huang et al. Citation2017).
The documented range of C. wuchihensis was recorded in Hainan, Yunnan, and Guangxi provinces in China, as well as in Vietnam (Wei et al. Citation2022). However, insights from Chen et al. (Citation2020) suggest a divergence in perspective. Specifically, they propose that the populations of C. wuchihensis_HN from Hainan and those of C. wuchihensis_GX from Guangxi and Vietnam are independent species, implying that the true C. wuchihensis could potentially be restricted to Hainan Island. Consequently, a comprehensive taxonomic evaluation of C. wuchihensis populations from Guangxi and Hainan is necessary. To contribute valuable molecular data regarding C. wuchihensis, we determined the mitochondrial genome sequence of C. wuchihensis from Hainan. The mitochondrial genome data of C. wuchihensis from Guangxi should be determined to resolve its taxonomic status. The completion of the mitochondrial genome sequence, determined in this study, will contribute to a better understanding of the conservation genetics and phylogenetics of this species.
Ethical approval
All procedures were approved by the Animal Research Ethics Committee of Hainan Provincial Education Center for Ecology and Environment, Hainan Normal University (HNECEE-2023-002).
Authors’ contributions
FT conceived the original idea. XH and YW carried out the experiment. FT analyzed the data and wrote the manuscript and all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (12.9 KB)Supplemental Material
Download MS Word (17.4 KB)Supplemental Material
Download MS Word (17.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OQ673175.
Extended data Figshare: the chromatogram view of mitochondrial genome of Crocidura wuchihensis (OQ673175). https://doi.org/10.6084/m9.figshare.23123255.v1
Additional information
Funding
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. doi:10.1093/nar/27.2.573.
- Chen SD, Chen D, Tang KY, Qin BX, Xie F, Fu CK, Liu Y, Liu SY. 2021. Discussion of taxonomic status of Crocidura dongyangjiangensis Liu Y, Chen SD and Liu SY, 2020 and Crocidura huangshanensis Yang, BW Zhang and Li, 2020. Acta Theriologica Sinica. 41(1):108–114.
- Chen SD, Qing J, Liu Z, Liu Y, Tang MK, Murphy RW, Pu YT, Wang XM, Tang KY, Guo KJ, et al. 2020. Multilocus phylogeny and cryptic diversity of white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in China. BMC Evol Biol. 20(1):29. doi:10.1186/s12862-020-1588-8.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi:10.1093/nar/gkz833.
- Huang ZH, Tu FY, Ke DH. 2017. Complete mitochondrial genome of Blue-throated Bee-eater Merops viridis (Coraciiformes: Meropidae) with its taxonomic consideration. PJZ. 49(1):79–84. doi:10.17582/journal.pjz/2017.49.1.79.84.
- Li H, Zhang G, Liu G, Sun X, Lin X, Li Y, Li Y. 2021. Characterization of the complete mitochondrial genome of Dongyangjiang White-toothed Shrew, Crocidura dongyangjiangensis (Eulipotyphla: Soricidae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(3):1004–1006. doi:10.1080/23802359.2021.1893617.
- Liu Y, Chen SD, Liu BQ, Liao R, Liu YX, Liu SY. 2020. A new species of the genus Crocidura (Eulipotyphla: soricidae) from Zhejiang Province, eastern China. Acta Theriologica Sinica. 40(1):1–12.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: a Laboratory Manual. (2nd ed). New York: Cold Spring Harbor Laboratory Press.
- Shaw TH, Wang S, Lu CK. 1966. A survey of the mammals of Hainan Island, China. Acta Zootaxonomica Sinica, 3(3):260–276.
- Smith AT, Xie Y, Wang S. 2009. A Guide to the Mammals of China. Changsha: Hunan Education Press.
- Song XH, Huang J, Yan CC, Xu GW, Zhang XY, Yue BS. 2015. The complete mitochondrial genome of Accipiter virgatus and evolutionary history of the pseudo-control regions in Falconiformes. Biochem Syst Ecol. 58:75–84. doi:10.1016/j.bse.2014.10.013.
- Swindell SR, Plasterer TN. 1997. SEQMAN. Sequence Data Analysis Guidebook. New York: Springer.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. doi:10.1093/molbev/mst197.
- Wei FW, Yang QS, Wu Y, Jiang XL, Liu SY. 2022. Taxonomy and distribution of mammals in China. Beijing: Science Press.
- Wilson DE, Reeder DM. 2005. Mammal species of the World: a taxonomic and geographic reference (3rd ed). Washington, DC:The Johns Hopkins University Press.