Abstract
Adonis pseudoamurensis W.T. Wang 1980 is an important traditional medicinal plant used for the treatment of cardiac diseases. The complete chloroplast (cp) genome of Adonis pseudoamurensis is reported for the first time in this study. The circular cp genome is 156,917 bp in length, consisting of a large single-copy region (86,262 bp), a small single-copy region (18,067 bp), and two inverted repeat regions (26,294 bp). The genome encodes 129 genes, comprising 84 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Phylogenetic analysis showed that A. pseudoamurensis is closely related to A. amurensis.
1. Introduction
The genus Adonis (Ranunculaceae) comprises 32 annual or perennial herbaceous species (Son, Citation2015). Owing to their complex chemical composition and cardiac-enhancing effects, species in Adonis are commonly used in folk medicine in Europe and China (Hao et al. Citation2015; Kaneko et al. Citation2008; Shang et al. Citation2019). Adonis pseudoamurensis W.T. Wang 1980 is an early spring flowering species mainly distributed in northeast China (Yang et al. Citation2015). This species is morphologically similar to A. amurensis but differs from it in having sessile cauline leaves and ovate to rhomboid sepals (Wang and Liu, Citation1988; Wang Citation1994a, Citation1994b). Based on internal transcriptional spacer sequence (ITS) and chloroplast sequences independent phylogenetic analyses of the genus Adonis confirmed that present Adonis taxa were genotypically differentiated according to their length of life and general morphology (Karahan et al. Citation2022; Seidl et al. Citation2022). A. pseudoamurensis has been synonymized with A. ramosa in Japan. However, similar morphological characteristics shared by the members of the genus Adonis render morphology-based identification of the species in the genus difficult (Suh et al. Citation2002). Therefore, it is necessary to distinguish A. pseudoamurensis from its closely related species in the genus Adonis with the help of molecular data. In this study, we sequenced the entire chloroplast genome of A. pseudoamurensis to provide useful information for the taxonomic and phylogenetic study of genus Adonis.
2. Materials and methods
Fresh leaves of wild A. pseudoamurensis were collected from the campus of Tonghua Normal University (41°44′33.10″N, 125°56′44.28″E) and stored in silica gel (). A voucher specimen was deposited in the Herbarium of Tonghua Normal University under the number sfxyyhsljcjzh20200429 (Liqiu Zhang, [email protected]). Total genomic DNA was extracted from silica-dried leaves using a modified CTAB method (Doyle and Doyle, Citation1987). DNA library preparation and paired-end read sequencing were performed on the Illumina NovaSeq 6000 platform. The cp genome was assembled using NOVOPlasty (NOVOPlasty4.2.pl -k) (Dierckxsens et al. Citation2017), and the A. sutchuenensis cp genome was used as the reference (Zhang et al. Citation2021). The depth of coverage was calculated by mapping the reads onto the chloroplast genome sequence with bowtie2 v2.3.4.3 (bowtie2 -x −1 -2 -S –sensitive-local -p 32) to determine the correctness of the assembly (Langmead and Salzberg, Citation2012). Chloroplast genome annotation was performed using the online web server CPGVAS2 (https://irscope.shinyapps.io/Chloroplot/) (Shi et al. Citation2019). The coding sites and codons of the predicted genes were manually corrected, and Geneious R11 (Biomatters Ltd., Auckland, New Zealand) was used to inspect the cp genome structure. The complete cp genome sequence was deposited in GenBank (accession number: MZ197990).
3. Result
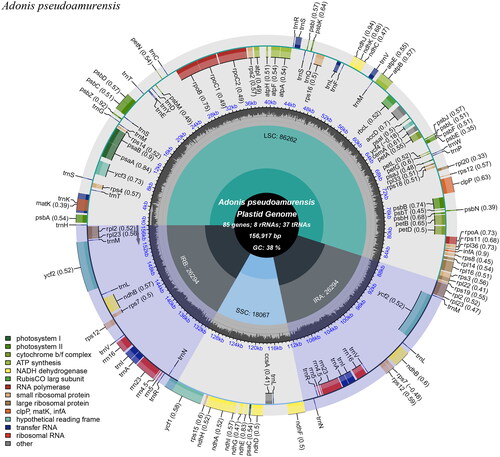
The complete cp genome of A. pseudoamurensis was 156,917 bp in length with an average depth of 337.62 X, and it included an 86,262 bp large single-copy region (LSC); an 18,067 bp small single-copy region (SSC); and two 26,294 bp inverted repeats (IRs) (Supplementary Figure 1). The GC content (%) of the whole genome, LSC, SSC, and IR regions was 37.95%, 36.14%, 31.55%, and 43.11%, respectively. A total of 129 genes were annotated, comprising 84 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Furthermore, 17 genes—seven tRNA, four rRNA, and six protein-coding genes—were repeated in the IR regions (). Gene structure analysis was done for ycf3, rpoC1, atpF, rps16, clpP, petB, petD, rpl16, rpl2, ndhB, ndhA difficult to annotate genes (Supplementary Figure 2). The LSC region contained 61 protein-coding and 22 tRNA genes, whereas the SSC region contained one tRNA gene and 11 protein-coding genes. Nineteen genes (11 protein-coding and eight tRNA genes) contained one intron, and two genes (ycf3 and clpP) had two introns. The rps12 gene is a trans-spliced gene, with the 5' end located in the LSC region and the copied 3' end located in the IR region.
Figure 2. Chloroplast genome map of A. pseudoamurensis. Genes drawn outside the outer circle are transcribed counterclockwise, and genes drawn inside the outer circle are transcribed clockwise. Genes belonging to different functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions.
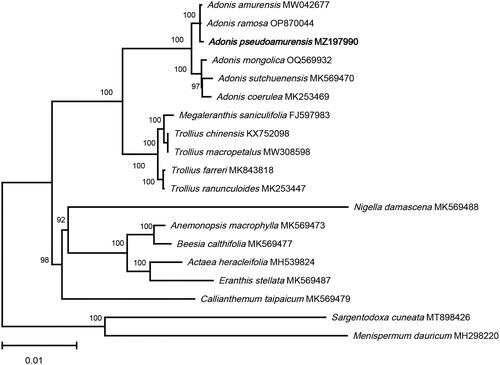
To elucidate the phylogenetic relationship between A. pseudoamurensis and other species of Ranunculaceae, 16 complete chloroplast genome sequences of Ranunculaceae were downloaded from NCBI, and the phylogenetic tree () was constructed using the maximum likelihood (ML) method implemented in FastTree 2.1.11 software (Price et al. Citation2009) with 1000 bootstrap replicates. Sargentodoxa cuneata (MT898426), and Menispermum dauricum (MH298220) were selected as outgroup species. In the ML tree, the four Adonis species are perennial species and clustered together in a well-supported clade (bootstrap support = 100%). A. pseudoamurensis and A. amurensis formed a highly supported clade (bootstrap support = 100%) that was sister to A. sutchuenensis, confirming the close phylogenetic relationship between A. pseudoamurensis and A. amurensis. The chloroplast genome sequence of A. pseudoamurensis provided here is a useful resource for further phylogenetic and taxonomic studies and species identification in the genus Adonis.
Figure 3. The ML phylogenetic tree is based on chloroplast genome sequences of Adonis pseudoamurensis species from the Ranunculaceae family, with Sargentodoxa cuneata (MT898426) and Menispermum dauricum (MH298220) as outgroups. Support values above the branches are ML bootstrap support. The following sequences were used: Trollius ranunculoides MK253447, Adonis coerulea MK253469 (He et al. Citation2019), Adonis pseudoamurensis MZ197990, Adonis ramosa OP870044, Megaleranthis saniculifolia FJ597983 (Kim et al. Citation2009), Trollius chinensis KX752098, Adonis sutchuenensis MK569470, Callianthemum taipaicum MK569479, Anemonopsis macrophylla MK569473, Beesia calthifolia MK569477, Eranthis stellata MK569487, Nigella damascena MK569488 (Zhai et al. Citation2019), Actaea heracleifolia MH539824 (Park et al. Citation2018), Menispermum dauricum MH298220 (Hina et al. Citation2018), Trollius farreri MK843818 (Yu et al. Citation2019), Adonis amurensis MW042677, Sargentodoxa cuneata MT898426 (Cui et al. Citation2021), Trollius macropetalus MW308598.
4. Discussion and conclusion
In this study, the chloroplast genome sequence of A. pseudoamurensis was assembled for the first time and the structure of this species was annotated. This phylogenetic result is similar to the phylogenetic analysis of the ribosomal ITS in A. pseudoamurensis (Suh et al. Citation2002). Comparison of the A. pseudoamurensis plastome to previously published data shows a high level of gene synteny with publicly available A. amurensis and A ramosa. This study provided useful information on the taxonomic and phylogenetic study of the genus Adonis.
Ethical approval
The material involved in the article does not involve ethical conflicts. This species is neither endangered on the CITES catalogue nor collected from a natural reserve, so it did not need specific permissions or licenses. All collection and sequencing work was strictly executed under local legislation and related laboratory regulations to protect wild resources.
Author contributions
The article was designed and conceived by XYZ and CSX; XYZ and LFZ assembled and annotated the cp genome; LQZ collected and identified the plant materia; XYZ contributed significantly to phylogenetic analysis and manuscript preparation; JYZ was involved in interpretation of the data and revised the manuscript critically for intellectual content. This article was modified and graphed by ZLZ. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (131.3 KB)Supplemental Material
Download TIFF Image (289.1 KB)Supplemental Material
Download TIFF Image (262 KB)Supplemental Material
Download MS Word (635.1 KB)Acknowledgement
The authors are grateful to the opened raw genome data from GenBank public database of National Center for Biotechnology Information (NCBI).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MZ197990. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA878453, SRR21533504, and SAMN30726550, respectively.
Additional information
Funding
References
- Cui X, Wang X, Wang Y, Yuan Q, Shen Y, Liu L. 2021. The complete chloroplast genome sequence of Sargentodoxa cuneata: genome structure and genomic resources. Mitochondrial DNA B Resour. 6(1):245–246. doi: 10.1080/23802359.2020.1863870.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hao DC, Xiao PG, Ma HY, Peng Y, He CN. 2015. Mining chemodiversity from biodiversity: pharmacophylogeny of medicinal plants of Ranunculaceae. Chin J Nat Med. 13(7):507–520. doi: 10.1016/S1875-5364(15)30045-5.
- He J, Yao M, Lyu RD, Lin LL, Liu HJ, Pei LY, Yan SX, Xie L, Cheng J. 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):15285. doi: 10.1038/s41598-019-51601-2.
- Hina F, Jin Z, Yang Z, Li P, Fu C. 2018. The complete chloroplast genome of Menispermum dauricum (Menispermaceae, Ranunculales). Mitochondrial DNA B Resour. 3(2):913–914. doi: 10.1080/23802359.2018.1501306.
- Karahan F, İlçim A, Türkoğlu A, İlhan E, Haliloğlu K. 2022. Phylogenetic relationship among taxa in the genus Adonis L. collected from Türkiye based on nrDNA internal transcribed spacer (ITS) markers. Mol Biol Rep. 49(8):7815–7826. doi: 10.1007/s11033-022-07607-7.
- Kaneko S, Nakagoshi N, Isagi Y. 2008. Origin of the endangered tetraploid Adonis ramosa (Ranunculaceae) assessed with chloroplast and nuclear DNA sequence data. Acta Phytotaxonomica et Geobotanica. 59(2):165–174.
- Kim YK, Park CW, Kim KJ. 2009. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol Cells. 27(3):365–381. doi: 10.1007/s10059-009-0047-6.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi: 10.1038/nmeth.1923.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Park I, Yang S, Kim WJ, Noh P, Lee HO, Moon BC. 2018. Complete chloroplast genome of Actaea heracleifolia (Kom.) J. Compton. Mitochondrial DNA B Resour. 3(2):939–940. doi: 10.1080/23802359.2018.1502636.
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. doi: 10.1093/molbev/msp077.
- Seidl A, Tremetsberger K, Pfanzelt S, Lindhuber L, Kropf M, Neuffer B, Blattner FR, Király G, Smirnov SV, Friesen N, et al. 2022. Genotyping-by-sequencing reveals range expansion of Adonis vernalis (Ranunculaceae) from Southeastern Europe into the zonal Euro-Siberian steppe. Sci Rep. 12(1):19074. doi: 10.1038/s41598-022-23542-w.
- Shang X, Miao X, Yang F, Wang C, Li B, Wang W, Pan H, Guo X, Zhang Y, Zhang J. 2019. The Genus Adonis as an Important Cardiac Folk Medicine: a Review of the Ethnobotany, Phytochemistry and Pharmacology. Front Pharmacol. 10:25. doi: 10.3389/fphar.2019.00025.
- Son D. 2015. A systematic study on the section Adonanthe of genus Adonis L. (Ranunculaceae) in East Asia. PhD thesis. Daejeon, Korea: Hannam University.
- Suh Y, Lee J, Lee S, Lee C, Yeau SH, Lee NS. 2002. Molecular evidence for the taxonomic identity of Korean Adonis (Ranunculaceae). J Plant Res. 115(3):217–223. doi: 10.1007/s102650200027.
- Wang LZ, Liu MY. 1988. Studies on the genus Adonis in northeast China. Bull Bot Res. 8(2):49–53.
- Wang WT. 1994a. Revision of Adonis (Ranunculaceae) (I). Bull Bot Res. 14:1–31.
- Wang WT. 1994b. Revision of Adonis (Ranunculaceae) (II). Bull Bot Res. 14:105–138.
- Yang WH, Zhang XW, Xu WJ, Huang HY, Ma YR, Bai H, Gong J, Ni SF. 2015. Overview of pharmacological research on Adonis l. Agric. Sci. Technol. 16(3):626–629.
- Yu B, Zhang R, Yang Q, Xu B, Liu ZL. 2019. The complete chloroplast genomes of Trollius farreri and Anemone taipaiensis (Ranunculaceae). Mitochondrial DNA B Resour. 4(2):3270–3271. doi: 10.1080/23802359.2019.1671248.
- Zhang LQ, Zhang XY, Hu YW, Sun RS, Yu JL. 2021. Complete chloroplast genome of Adonis amurensis (Ranunculaceae), an important cardiac folk medicinal plant in east Asia. Mitochondrial DNA B Resour. 6(2):583–585. doi: 10.1080/23802359.2021.1875916.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21. doi: 10.1016/j.ympev.2019.02.024.

