Abstract
As one of the top 10 famous flowers in Chinese tradition, Rhododendron przewalskii subsp. przewalskii known as ‘beauty in flowers,’ which has high ornamental and medicinal value. The complete chloroplast (cp) genome of R. przewalskii subsp. przewalskii was determined in this study. The complete chloroplast genome of R. przewalskii subsp. przewalskii was 201,233 bp in length and contained a large single-copy region (LSC, 108,077 bp), and a small single-copy region (SSC, 2624 bp) and a pair of inverted repeat regions (IRa and IRb, 45,266 bp). A total of 142 functional genes were observed in this cp genome, including 91 protein-coding genes (PCGs), 43 transfer RNA genes (tRNAs), and eight ribosomal RNA genes (rRNAs). The R. przewalskii subsp. przewalskii cp genome has an A + T content of 64.06% and presents a positive AT-skew (0.53%) and a negative GC-skew (−1.56%). The maximum likelihood phylogenetic analysis based on the concatenated nucleotide sequences of 13 PCGs strongly supported the monophyletic relationship of R. przewalskii subsp. przewalskii the clade of R. henanense subsp. lingbaoense. This study provides genomic evidence for the vegetation classification of Rhododendron.
Introduction
The Rhododendron przewalskii subsp. przewalskii Maximowicz (Maximowicz 1877) belongs to the genus Rhododendron, which was discovered and named by Maximowicz in 1877. Its long leaves and flowers are used as medicine for treating peliosis and other diseases, and is one of the ethnically distinctive medicinal herbs in Tibetan areas of China (Luo Citation2018). R. przewalskii subsp. przewalskii is reported to contain mainly knotweed glycosides, cottonphenol-3-O-β-D-galactosidase, quercetin, birch glucoside, β-sitosterol and other major chemical components, which are widely used in Chinese medicine, proprietary Chinese medicine and compound medicine for clinical treatment of cough and pneumonia (Dai et al. Citation2021). Rhododendron M. is the largest and most diverse genus in the family, with more than 1000 species worldwide and about 571 species in China, distributed in southwestern and southern China. The genus Rhododendron is internationally renowned for its wide variety of species, beautiful flowers, and lustrous colors, and is known as the ‘king of woody flowers’ and the ‘beauty of flowers’. China has the largest number of Rhododendron resources in the world, but research on the domestication, breeding, and exploitation of germplasm resources of Rhododendron is lagging far behind that of other countries with developed garden industries (Zhuang Citation2019). It is easy to find that this is mainly due to two reasons, the first lies in the strongly differentiated radiation of Rhododendron and the frequent interspecific hybridization of reproduction (Ma et al. Citation2016), and the second is the late start and weak foundation of Rhododendron breeding and utilization in China, which limits the rational utilization of Rhododendron resources (Liu et al. Citation2021). Further research on this species is therefore of great medicinal and economic value.
Chloroplast genes provide reliable scientific genetic information for plant taxonomy, phylogeny, and species evolution, and respond to the origin and migration of species (Tang et al. Citation2022). Most higher plant chloroplast genomes are maternally inherited, structurally conserved, and contain a large amount of genetic information, which is a common source of information for plant genetic diversity analysis, adaptive evolution, and molecular breeding studies (Zhang et al. Citation2022). However, the existing classification of the genus Rhododendron is limited to taxonomic units, such as genera, subgenera, and groups, while the classification of subspecies, species, and subgroups is neglected, which is also caused by the lack of information on more chloroplast genomes of Rhododendron.
In this study, we analyze the sequence and structural features of Rhododendron subspecies based on the sequencing, assembly, and annotation of Rhododendron subspecies my research material, and analyze its evolutionary developmental position in the genus Rhododendron, which will provide genetic resources for genetic breeding, species identification and resource development of Rhododendron subspecies and Rhododendron genus.
Materials and methods
Sample collection, DNA extraction, chloroplast genome sequencing, and assembly
In June 2022, the author collected fresh samples of the whole plant of R. przewalskii subsp. przewalskii () in Xiazhasha Village, Xunhua County, Haidong City, Qinghai Province, China (102°44′8″E, 35°34′ 23″N, Altitude: 3499 m). The samples have been deposited in the Herbarium of Qinghai College of Animal Husbandry and Veterinary Medicine, Qinghai University, Xining City, Qinghai Province, China (LSDJ-cp17, Xiuzhang Li, [email protected]).
Figure 1. Morphological characteristics of R. przewalskii subsp. przewalskii. The nutritional growth stage of R. przewalskii subsp. przewalskii, the leaf blade is leathery, often set at the end of the branch in an ovate-elliptic shape, and the apex is obtuse but with a small acumen. Its base is rounded or slightly heart-shaped, dark green above, glabrous, slightly wrinkled, midrib concave, lateral veins 11–12 pairs, retuse. In June 2022, the author took this photo in Xiazhasha Village, Xunhua County, Haidong City, Qinghai Province, China (102°44′8″E, 35°34′ 23″N, Altitude: 3499 m).

The library was prepared using the Vazyme ND627 kit with a 1ug gDNA template. The size of the inserted fragment of the library was 200–400 bp. After the construction of the library, Qubit3.0 was used for preliminary quantification, and Agilent 2100 was used to detect the insert size of the library. Finally, the effective concentration of the library was accurately quantified by Q-PCR. The qualified library was tested and a double-ended sequencing program (PE150) was run on NovaSeq 6000 sequencing platform, and 150 bp double-ended reads were obtained. For Oxford Nanopore sequencing, the libraries were prepared using the SQKLSK109 ligation kit and using the standard protocol. The purified library was loaded onto primed R9.4 Spot-On Flow Cells and sequenced using a PromethION sequencer (Oxford Nanopore Technologies, Oxford, UK). Base calling analysis of raw data was performed using the Oxford Nanopore GUPPY software.
Chloroplast genome annotation and characterization
Chloroplast genome annotation was performed using GeSeq software (Bolger et al. Citation2014), and annotation results were manually corrected using Geneious Prime (Kearse et al. Citation2012). The genome map was drawn using the Organellar Genome DRAW tool v.1.3.1 for further comparison of gene order and content.
Phylogenomic analysis
To accurately determine the phylogenetic position of R. przewalskii subsp. przewalskii in Rhododendron, the authors collected 18 species from NCBI (https://www.ncbi.nlm.nih.gov) for the phylogenetic tree in their analysis. Species were selected as follows: among the five genera belonging to the Ericaceae J., at least one species from each genus was selected, and if there were more species in the genus then up to three species that have been published in NCBI were selected for the phylogenetic tree construction, and Plantago asiatica of Plantago L. was used as an outgroup. The chloroplast genomes of the above 19 species were analyzed using the maximum likelihood tree method (ML) in RAxML 8.2.10 (Stamatakis Citation2014) to obtain bootstrap values and support for each branch of the developmental tree at Bootstrap = 1000 and generate a phylogenetic tree (Kozlov et al. Citation2019). The following sequences were used: Rhododendron przewalskii subsp. przewalskii, OQ054014 (this study), Gaultheria hookeri NC071361 (Li et al. Citation2021), Gaultheria nummularioides MZ457321 (Zhang et al. Citation2022), Gaultheria praticola KY652194 (Zhang et al. Citation2022), Gaultheria griffithiana OM048893 (Li et al. Citation2021), Gaultheria fragrantissima NC059849 (Xu et al. Citation2021), Agapetes malipoensis NC058759 (Huong and Hiep Citation2012), Vaccinium bracteatum LC521967 (Kim et al. Citation2022), Vaccinium oldhamii NC042713 (Kim et al. Citation2019), Vaccinium uliginosum LC521968 (Hong et al. Citation2019), Vaccinium macrocarpon JQ248601 (Hong et al. Citation2019), Rhododendron griersonianum NC050162 (Liu, Fu, et al. Citation2020), Rhododendron delavayi NC047438 (Liu, Chen, et al. Citation2020), Rhododendron henanense subsp. lingbaoense MT239363 (Zhou et al. Citation2021), Rhododendron calophytum NC061396 (Ma et al. Citation2022), Rhododendron huadingense NC063113 (An et al. Citation2022), Arbutus unedo JQ067650 (Fernando et al. Citation2013), and Plantago asiatica NC060521 (Si et al. Citation2022).
Results and discussion
Structure and function of the chloroplast genome of R. przewalskii subsp. przewalskii
The authors verified the reliability of the determination of the chloroplast genome of R. przewalskii subsp. przewalskii by annotating it with an online tool (http://www.1kmpg.cn/cpgview/). The results showed that the authors’ sequencing depth was above 15X (Supplementary Figure 2(A)), indicating that both the measured genome coverage (Supplementary Figure 2(B)) and sequencing error rate control were guaranteed, and the sequencing results and annotation results were accurate.
The complete chloroplast genome of R. przewalskii subsp. przewalskii was assembled and spliced into a covalently closed loop tetrameric structure with a total sequence length of 201,233 bp, containing an LSC region of 108,077 bp, an SSC region of 2624 bp and a pair of reverse repeat IR (IRa and IRb) regions of 45,266 bp (). The GC content in its chloroplast whole genome was 35.94, 35.32, 36.86, and 29.88% in the LSC, IR, and SSC regions, respectively. species should theoretically have a balanced probability of base substitution under natural selection pressure, however, its chloroplast genome had AT-skew = 0.53% and GC-skew = −1.56%, which suggests that under natural selection pressure, its G + C bases tend to be replaced by A + T.
Figure 2. Annotated map of the circular chloroplast genome of R. przewalskii subsp. przewalskii. The genes inside the circles are transcribed clockwise and those outside are transcribed counterclockwise. The dark gray in the inner circle shows GC content, while the light gray shows a + T content (the dashed area in the inner circle indicates the content of the GC chloroplast genome). The legend identifies genes with different functions.
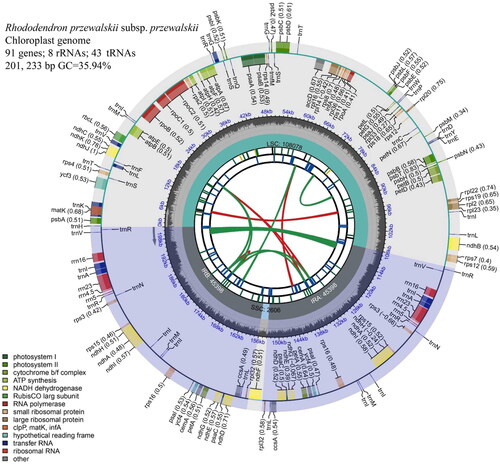
A total of 142 functional genes were observed in its chloroplast genome, including 91 protein-coding genes, 43 tRNA genes, and 8 rRNA genes. These 91 PCGs consisted of 57 different PCGs, including 12 double-copy genes (rpl32, ycf4, ycf1, cemA, ccsA, psaC, psaI, ndhA, ndhE, ndhI, rps16, and rps18), two three-copy genes (rps3 and rps19) and one four-copy gene (ndhG). Among them, 60 PCGs were distributed in the LSC region, 30 PCGs were distributed in the IR region, and one PCG (ndhG) was distributed in the SSC region. Among the 12 double-copy PCGs, two copies of rps18 were distributed in the LSC region and the other double-copy genes were in the IR region. Among the two three-copy PCGs, rps3 appeared for the first time in the LSC region and then repeated twice in the IR region, and rps19 was similar. A four-copy gene ndhG appears only in the IR region.
These 43 tRNA genes consisted of 27 different tRNA genes, including 7 double-copy genes (rnV-GAC, trnA-UGC, trnI-GAU, trnL-UAG, trnN-GUU, trnQ-UUG, and trnR-ACG), 2 three-copy genes (trnM-CAU and trnT-UGU) and 1 six copy genes (trnI-CAU), of which 24 tRNA genes were distributed in the LSC region, 18 tRNA genes were distributed in the IR region, and only one tRNA gene (trnL-UAG) was in the SSC region. Among the seven double-copy tRNA genes, both repeats of trnQ-UUG were distributed in the LSC region, and two repeats of trnL-UAG appeared adjacent to each other, the former at the end of the IRA region and the latter in the SSC region, the only tRNA gene encoded by the SSC region, while the other double-copy genes were distributed in the IR region. The three copies of trnM-CAU appeared first in the LSC region and then repeated twice in the IR region, and trnT-UGU was similar. The first two repeats of the six copies of trnI-CAU were distributed in the LSC region and the last four repeats were distributed in the IR region. The eight rRNA genes included four different rRNA genes (rrn4.5S, rrn5S, rrn16S, and rrn23S), which are all double-copy genes and are distributed in the IR region.
Phylogenomic analysis
In this study, we reconstructed the phylogenetic status of R. przewalskii subsp. przewalskii based on 18 reported species of Rhododendron, and the phylogenetic results showed that 19 species were clustered into three branches, and R. przewalskii subsp. przewalskii was closest to R. henanense subsp. lingbaoense strong bootstrap value of 1000 ().
Figure 3. Maximum likelihood tree based on the chloroplast PCGs sequences of R. przewalskii subsp. przewalskii and 18 species of Ericaceae. The numbers shown next to the nodes are bootstrap support values and GenBank accession numbers of each species are listed in the tree. Red font for this study. Numbers >1 in the plot indicate support for species relatedness, and numbers <1 are bootstrap values, indicating node confidence levels.

Conclusion
The genus Rhododendron is rich in species and is both a medicinal and ornamental flower, thus it has received much attention from scholars, and its interspecific evolutionary relationships are one of the most relevant hot issues for scholars. However, the species of Rhododendron are strongly differentiated, and more genetic evidence is needed to analyze the interspecific evolutionary relationships. In this study, we have completed the determination and assembly of the complete chloroplast genes of R. przewalskii subsp. przewalskii for the first time and annotated them accordingly, which strengthens the current understanding of the evolutionary relationships of Rhododendron species and provides genetic evidence to further elucidate the phylogenetic relationships of the Rhododendron genus.
Ethical approval
This study did not involve endangered or protected species, and the sampling sites were not in any protected areas. The collection of plant materials was in line with the regulations of the Qinghai Provincial Government and was approved by the Xunhua County Government of Haidong City.
Author contributions
Tao Wang conceived, designed, and completed the manuscript. Yuling Li and Xiuzhang Li performed field collection and identification of plant material. Zhengfei Cao and Hui He assembled and annotated the chloroplast genome lineage sequences. Chuyu Tang and Jianbo Chen performed a phylogenetic analysis and visualized the maps. All authors discussed the results, critiqued the manuscript, and validated the current version of the manuscript.
Supplemental Material
Download MS Word (2.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Supplementary Figure 2(A) is the sequencing depth map and coverage map and Supplementary Figure 2(B) is the black-white arrow shows the cis-spliced genes (left), and the black-grey-white arrow shows trans-spliced genes (rps12) (right), which the authors have placed in the Supplementary.
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under the accession(https://www.ncbi.nlm.nih.gov/nuccore/OQ054014.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA947480, SRR23938567, and SAMN33850058, respectively.
Additional information
Funding
References
- An R, Niu MY, Lou XZ, Huang HL, Lin E. 2022. The complete chloroplast genome of Rhododendron huadingense (Ericaceae). Mitochondrial DNA B Resour. 7(11):1910–1912. doi: 10.1080/23802359.2022.2135403.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Dai L, He J, Miao X, Guo X, Shang X, Wang W, Li B, Wang Y, Pan H, Zhang J. 2021. Multiple biological activities of Rhododendron przewalskii Maxim. extracts and UPLC-ESI-Q-TOF/MS characterization of their phytochemical composition. Front Pharmacol. 12:599778. doi: 10.3389/fphar.2021.599778.
- Fernando MA, Del Campo EM, Lázaro-Gimeno D, Mezquita-Claramonte S, Molins A, Mateu-Andrés I, Pedrola-Monfort J, Casano LM, Barreno E. 2013. Balanced gene losses, duplications, and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLOS One. 8(11):e79685. doi: 10.1371/journal.pone.0079685.
- Hong S, Cheon K, Yoo K, Lee H, Mekapogu M, Cho K. 2019. Comparative analysis of the complete chloroplast genome sequences of three Amaranthus species. Plant Genet Resour. 17(3):245–254. doi: 10.1017/S1479262118000485.
- Huong NT, Hiep NT. 2012. Agapetes malipoensis S. H. Huang (Ericaceae juss.), a new species for the flora of Vietnam. V J Bio. 32(1):e649. doi: 10.15625/0866-7160/v32n1.649.
- Kearse M, Moir R, Wilson A, Steven SH, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kim BY, Kwon EB, Yang HJ, Li W, Hwang YH, Kim YS, Pak ME, Go YH, Choi JG. 2022. Vaccinium bracteatum Thunb extract inhibits HSV-1 infection by regulating ER stress and apoptosis. Antioxidants. 11(9):1773. doi: 10.3390/antiox11091773.
- Kim SC, Baek SH, Lee JW, Hyun HJ. 2019. Complete chloroplast genome of Vaccinium oldhamii and phylogenetic analysis. Mitochondrial DNA Part B. 4(1):902–903. doi: 10.1080/23802359.2019.1579067.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi: 10.1093/bioinformatics/btz305.
- Li YR, Xu YL, Du XY, Yang SD, Lu L. 2021. Characterization of the complete plastid genome of Gaultheria griffithiana (Ericaceae). Mitochondrial DNA B Resour. 6(5):1575–1577. doi: 10.1080/23802359.2021.1914227.
- Liu DT, Fu CN, Yin LJ, Ma YP. 2020. Complete plastid genome of Rhododendron griersonianum, a critically endangered plant with extremely small populations (PSESP) from southwest China. Mitochondrial DNA B Resour. 5(3):3086–3087. doi: 10.1080/23802359.2020.1800427.
- Liu J, Chen T, Zhang YB, Li YK, Gong JY, Yi Y. 2020. The complete chloroplast genome of Rhododendron delavayi (Ericaceae). Mitochondrial DNA B Resour. 5(1):37–38. doi: 10.1080/23802359.2019.1689860.
- Liu N, Lu Z, Zhou YL, Tu ML, Wu ZZ, Gui DP, Ma YP, Wang JH, Zhang CJ. 2021. The Rhododendron plant genome database (RPGD): a comprehensive online omics database for Rhododendron. BMC Genomics. 22(1):376. doi: 10.1186/s12864-021-07704-0.
- Luo DS. 2018. The original interpretation of Jingzhu Materia Medica. Chengdu: Sichuan Science and Technology Press; p. 485.
- Ma WB, Yu T, Wang WJ, Wang F, Ji HJ, Mu CL. 2022. The complete chloroplast genome of Rhododendron calophytum Franch (Ericaceae). Mitochondrial DNA Part B. 7(9):1758–1759. doi: 10.1080/23802359.2022.2106161.
- Ma YP, Xie WJ, Sun WB, Marczewski T. 2016. Strong reproductive isolation despite occasional hybridization between a widely distributed and a narrow endemic Rhododendron species. Sci Rep. 6(1):1–11.
- Si H, Li R, Zhang QD, Liu LX. 2022. Complete chloroplast genome of Plantago asiatica and its phylogenetic position in Plantaginaceae. Mitochondrial DNA B Resour. 7(5):819–821. doi: 10.1080/23802359.2022.2073838.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Tang CY, Li XZ, Chen JB, Liang J, Wang T, Yuling L. 2022. Characterization and phylogenetic relationship of the complete chloroplast genome of a Chinese traditional medicinal plant Potentilla anserina. Mitochondrial DNA B Resour. 7(9):1653–1655. doi: 10.1080/23802359.2022.2119817.
- Xu YL, Du X-Y, Yi-Rong L, Lu L. 2021. The complete chloroplast genome of Gaultheria fragrantissima Wall. (Ericaceae) from Yunnan, China, an aromatic medicinal plant in the winter greens. Mitochondrial DNA B Resour. 6(6):1761–1762. doi: 10.1080/23802359.2021.1923425.
- Zhang J, Cheng XJ, Fritsch Peter W, Li Y, Yang SD, Lu L. 2022. Genetic variation in Gaultheria nummularioides (Ericaceae: Gaultherieae) from the Sky Islands of the Himalaya-Hengduan Mountains. Plant Diversity. 14(8):e652. doi: 10.3390/d14080652.
- Zhang YM, Han LJ, Yang CW, Yin ZL, Tian Y, Qian ZG, Li DG. 2022. Comparative chloroplast genome analysis of medicinally important Veratrum (Melanthiaceae) in China: insights into genomic characterization and phylogenetic relationships. Plant Divers. 44(1):70–82. doi: 10.1016/j.pld.2021.05.004.
- Zhou XJ, Wei MJ, Zhang K, Han JW, Wang HL, Dong SW. 2021. Characterization of the complete chloroplast genome of Rhododendron henanense subsp. lingbaoense Fang. Mitochondrial DNA B Resour. 6(12):3325–3326. doi: 10.1080/23802359.2021.1994898.
- Zhuang P. 2019. Progress on the fertility of Rhododendron. Biodivers Sci. 27(3):327–338.
