Abstract
Jatropha curcas (Linnaeus, 1753) is a plant species in the order Malpighiales and the family Euphorbiaceae and is native to the tropical regions of America, such as Mexico and Argentina. Currently, this plant species inhabits tropical and subtropical regions of the world. Jatropha has been widely used as a biofuel plant to produce high-quality diesel engine fuel. In this study, the complete mitochondrial genome sequence of J. curcas was assembled into 561,839 bp circular nucleotides with a GC content of 44.6%. The mitochondrial genome of J. curcas comprises 33 known protein-coding genes, 22 tRNA genes, three rRNA genes, one ncRNA gene, and 85 open reading frame genes. Phylogenetic analysis showed this species is closely related to the castor bean (Ricinus communis).
Introduction
Jatropha curcas (Linnaeus, 1753) is a monoecious tree or shrub that originated in Central America and is widely distributed in tropical and subtropical regions. J. curcas belongs to the order Malpighiales and the family Euphorbiaceae. Euphorbiaceae is one of the prominent flowering plant families, including several economically important plant species such as, rubber trees (Hevea brasiliensis), cassava (Manihot esculenta), and castor bean (Ricinus communis) (Ha et al. Citation2019; Fayed et al. Citation2020). As a member of the Euphorbiaceae, J. curcas has been widely used as a biofuel plant to generate high-quality diesel fuel because it produces seeds that contain significant portions of non-edible oil (Ashraful et al. Citation2014; Takase et al. Citation2015). Jatropha seed oil contains oleic (18:1), linoleic (18:2), palmitic (16:0), and stearic acid (18:0), along with phorbol ester, a toxic compound, suggesting that it is a promising alternative to standard diesel fuel that does not compete for food security (Sinha et al. Citation2015; Gomes et al. Citation2022). Furthermore, the evolutionarily conserved succulent stem in Jatropha and some Euphorbiaceae plants makes Jatropha tolerant to drought stress and arid lands, facilitating marginal land use and reducing competition for arable land use in food cultivation (Maes et al. Citation2009; Santos et al. Citation2015; Della Torre et al. Citation2021).
The complete chloroplast genome sequence has played a crucial role in understanding the evolutionary relationships of various Euphorbiaceae plants (Zhang, Shi, et al. Citation2019; Zhang, Zhao, et al. Citation2019; Wang et al. Citation2020; Iwata et al. Citation2022). The whole reference genome sequences and complete circular chloroplast genome sequence of J. curcas have been reported recently, providing insights into its evolutionary history (Asif et al. Citation2010; Ha et al. Citation2019). However, the complete mitochondrial genome sequence has not been reported in Euphorbiaceae, except Ricinus communis (Rivarola et al. Citation2011). Here, we report the novel complete mitochondrial genome sequence of J. curcas as a second representative mitochondrial genome sequence in Euphorbiaceae plants.
Materials and methods
A specimen of J. curcas was determined by D. Burch and deposited at the Atlas of Florida Plants (https://florida.plantatlas.usf.edu/Default.aspx, Richard P. Wunderlin, [email protected]) under the accession number: 106202 (). The whole-genome sequence of J. curcas var. Chai Nat, widely cultivated in Chai Nat province (latitude 15.18567 and longitude 100.12367) of Thailand, has been reported previously (Ha et al. Citation2019) (). To investigate the mitochondrial genome sequence of J. curcas, we downloaded raw PacBio read sequences (SRR5974849) and Illumina paired-end sequences (SRR5974850) from PRJNA399212 of SRA (Ha et al. Citation2019). To assemble the mitochondrial genome, we corrected and trimmed the PacBio reads using Canu (Koren et al. Citation2017). Sequence reads carrying homologous sequences to the mitochondrial genes of Ricinus communis (NC_015141.1) were selected using TBLASTX (Camacho et al. Citation2009). The selected reads were initially assembled using FALCON (Chin et al. Citation2016). A total of nine contigs carrying the mitochondrial genes were identified. The assembled contigs were submitted as seed sequences to the short read assembly using NOVOPlasty (Dierckxsens et al. Citation2017). The assembled sequence was assessed by coverage and sequencing depth (https://www.protocols.io/view/generating-sequencing-depth-and-coverage-map-for-o-4r3l27jkxg1y/v1) (Supplemental figure 1). To validate the complete circular form of J. curcas mitogenome, sequence alignment and mapping depth around the merged position were examined using Samtools and IGV (Li et al. Citation2009; Thorvaldsdóttir et al. Citation2013) (Supplemental figure 2). Gene prediction and annotation were performed using MFannot (https://megasun.bch.umontreal.ca/apps/mfannot/) for protein-coding genes and transfer RNAs and GeSeq annotator for ribosomal RNAs (Tillich et al. Citation2017). A map of the complete mitochondrial genome sequence and genes was generated using OrganellarGenomeDRAW (Greiner et al. Citation2019). A molecular phylogenetic tree was constructed using the maximum-likelihood method with Jones–Taylor–Thornton (JTT) matrix-based model and 1000 bootstrap replications using MEGA software (Kumar et al. Citation2018).
Figure 1. The specimen and picture of the Jatropha curcas L. The specimen of J. curcas was deposited at the Atlas of Florida Plants, operated by the Institute for Systematic Botany at the University of South Florida (https://Florida.plantatlas.usf.edu/Default.aspx) under the accession number: 106202 (A). Picture of J. curcas was taken and provided by Won Joo Hwang (B).

Results
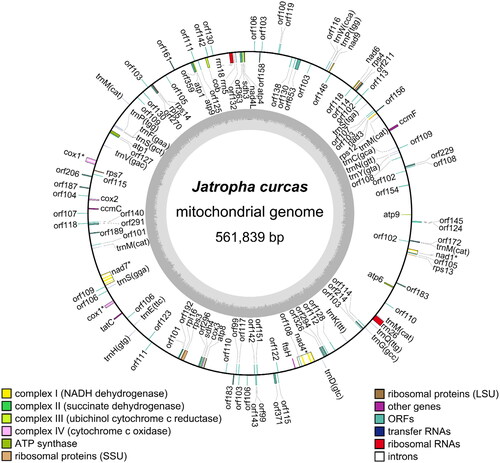
The complete mitochondrial genome of J. curcas spans 561,839 bp in length with 44.6% GC content and encodes 144 genes, including 33 known protein-coding genes, 22 tRNA genes, three rRNA genes, one ncRNA gene, and 85 open reading frame genes (ORFs) (). Among the 33 known protein-coding genes, nad1 and cox1 (first copy) carried two exons, and nad4 and nad7 carried 4 and 5 exons, respectively (Supplemental figure 3). There were 15 duplicated gene pairs, including atp1, atp9, cox1, trnP, and 11 orf genes (orf99, orf101, orf105, orf107, orf110, orf111, orf114, orf115, orf118, orf142, and orf183), and four sets of triplicated orf genes (orf102, orf108, orf109, and orf130). Interestingly, two ORF genes (orf106 and orf103) were quadruplicated and hexaplicated, respectively. A set of trnM genes was presented as pentaplicated genes. The remaining 87 genes are presented as single copies.
Figure 2. Map of Jatropha curcas mitochondrial genome. The genes annotated outside the circular genome are in the forward orientation, whereas the genes inside the circle are in the reverse orientation. Genes with asterisk mark indicate cis-splicing genes.
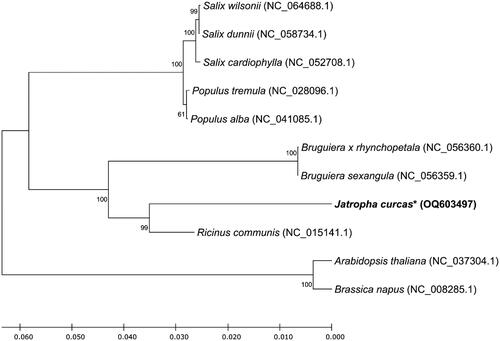
A total 18 genes, including atp1 (second copy), atp4, atp6, ccmC, ccmF, cob, cox1 (first copy), cox2, cox3, nad4, nad6, nad7, nad9, orf206, orf294, orf326, orf653, and rps3 have substantially conserved over 11 plant species including nine Malpighiales plants and two outgroup plants: Arabidopsis thaliana (Sloan et al. Citation2018), Brassica napus (Handa Citation2003), Bruguiera sexangula (Zhang, Bai, Zhang Citation2020), Bruguiera x rhynchopetala (Zhang, Bai, Liu Citation2020), Populus alba (Brenner et al. Citation2019), Populus tremula (Kersten et al. Citation2016), Ricinus communis (Rivarola et al. Citation2011), Salix cardiophylla (Chen et al. Citation2020), Salix dunnii (He et al. Citation2021), and Salix wilsonii (Han et al. Citation2022). The phylogenetic tree showed that J. curcas was closely related to R. communis ().
Figure 3. Unrooted phylogenetic tree of mitochondrial genome sequences of Malpighiales. The phylogenetic tree was constructed using the maximum-likelihood (ML) method with Jones–Taylor–Thornton (JTT) matrix-based model and 1000 times bootstrap replications. The protein sequences of mitochondrial genes from nine Malpighiales and two outgroup plants (Arabidopsis thaliana and Brassica napus) were used. Bootstrap values indicate the topology of the tree. The asterisk mark indicates the plant species investigated in this study.
Discussion and conclusions
We report the novel complete mitochondrial genome sequence of J. curcas as a second representative plant species in the Euphorbiaceae family. The complete mitochondrial genome sequence of J. curcas and its whole genome sequences provide an essential resource for phylogenomic analysis and will help elucidate the evolutionary relationship between the Malpighiales and the Euphorbiaceae family soon.
Author contributions
S.S. and J.H. conceptualized the study and conducted bioinformatics analyses. The manuscript was prepared by S.S. and revised by J.H.
Ethical approval
Any data from NCBI SRA does not need ethical approval.
Supplemental Material
Download PDF (288.4 KB)Supplemental Material
Download MS Word (1.5 MB)Acknowledgements
We especially thank Won Joo Hwang for providing a picture of Jatropha curcas.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ603497. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA399212, SRR5974849, SRR5974850, and SAMN07527279, respectively.
Additional information
Funding
References
- Ashraful AM, Masjuki HH, Kalam MA, Rizwanul Fattah IM, Imtenan S, Shahir SA, Mobarak HM. 2014. Production and comparison of fuel properties, engine performance, and emission characteristics of biodiesel from various non-edible vegetable oils: a review. Energy Convers Manage. 80:202–228. doi: 10.1016/j.enconman.2014.01.037.
- Asif MH, Mantri SS, Sharma A, Srivastava A, Trivedi I, Gupta P, Mohanty CS, Sawant SV, Tuli R. 2010. Complete sequence and organisation of the Jatropha curcas (Euphorbiaceae) chloroplast genome. Tree Genet Genomes. 6(6):941–952. doi: 10.1007/s11295-010-0303-0.
- Brenner WG, Mader M, Müller NA, Hoenicka H, Schroeder H, Zorn I, Fladung M, Kersten B. 2019. High level of conservation of mitochondrial RNA editing sites among four Populus species. G3. 9(3):709–717. doi: 10.1534/g3.118.200763.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10(1):421. doi: 10.1186/1471-2105-10-421.
- Chen X, Zhang L, Huang Y, Zhao F. 2020. Mitochondrial genome of Salix cardiophylla and its implications for infrageneric division of the genus of Salix. Mitochondrial DNA B Resour. 5(3):3485–3486. doi: 10.1080/23802359.2020.1827065.
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single molecule real-time sequencing. Nat Methods. 13(12):1050–1054. doi: 10.1038/nmeth.4035.
- Della Torre F, Ferreira BG, Lima JE, Lemos-Filho JP, Rossiello ROP, França MGC. 2021. Leaf morphophysiological changes induced by long-term drought in Jatropha curcas plants explain the resilience to extreme drought. J Arid Environ. 185:104381. doi: 10.1016/j.jaridenv.2020.104381.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Fayed A, Soliman M, Faried A, Hassan M. 2020. Taxonomic evaluation of Euphorbiaceae sensu lato with special reference to Phyllanthaceae as a new family to the Flora of Egypt. Biol Forum. 11:47–64.
- Gomes TG, Isa Abdel Hadi SI, Antônio de Aquino Ribeiro J, Segatto R, Mendes TD, Helm CV, Chagas Júnior AF, Gerard Miller RN, Mendonça S, Gonçalves de Siqueira F. 2022. Phorbol ester biodegradation in Jatropha curcas cake and potential as a substrate for enzyme and Pleurotus pulmonarius edible mushroom production. Biocatal Agric Biotechnol. 45:102498. doi: 10.1016/j.bcab.2022.102498.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Ha J, Shim S, Lee T, Kang YJ, Hwang WJ, Jeong H, Laosatit K, Lee J, Kim SK, Satyawan D, et al. 2019. Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol J. 17(2):517–530. doi: 10.1111/pbi.12995.
- Han F, Qu Y, Chen Y, Xu L, Bi C. 2022. Assembly and comparative analysis of the complete mitochondrial genome of Salix wilsonii using PacBio HiFi sequencing. Front Plant Sci. 13:1031769. doi: 10.3389/fpls.2022.1031769.
- Handa H. 2003. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31(20):5907–5916. doi: 10.1093/nar/gkg795.
- He L, Jia K, Zhang R, Wang Y, Shi T, Li Z, Zeng S, Cai X, Wagner ND, Hörandl E, et al. 2021. Chromosome‐scale assembly of the genome of Salix dunnii reveals a male‐heterogametic sex determination system on chromosome 7. Mol Ecol Resour. 21(6):1966–1982. doi: 10.1111/1755-0998.13362.
- Iwata H, Ito T, Kokubugata G, Takayama K. 2022. The complete chloroplast genome of a coastal plant, Euphorbia jolkinii (Euphorbiaceae). Mitochondrial DNA B Resour. 7(3):569–570. doi: 10.1080/23802359.2022.2055502.
- Kersten B, Rampant PF, Mader M, Paslier M-CL, Bounon R, Berard A, Vettori C, Schroeder H, Leplé J-C, Fladung M. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLOS One. 11(1):e0147209. doi: 10.1371/journal.pone.0147209.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352.
- Maes WH, Achten WMJ, Reubens B, Raes D, Samson R, Muys B. 2009. Plant–water relationships and growth strategies of Jatropha curcas L. seedlings under different levels of drought stress. J Arid Environ. 73(10):877–884. doi: 10.1016/j.jaridenv.2009.04.013.
- Rivarola M, Foster JT, Chan AP, Williams AL, Rice DW, Liu X, Melake-Berhan A, Creasy HH, Puiu D, Rosovitz MJ, et al. 2011. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLOS One. 6(7):e21743. doi: 10.1371/journal.pone.0021743.
- Santos MW, dos Silva CF, da França MGC, Zonta E, Rossiello ROP. 2015. The relationship between succulence and shoot biomass differences according to nutritional status in Jatropha curcas L. AJAR. 10:4025–4031.
- Sinha P, Islam MA, Negi MS, Tripathi SB. 2015. Changes in oil content and fatty acid composition in Jatropha curcas during seed development. Ind Crops Prod. 77:508–510. doi: 10.1016/j.indcrop.2015.09.025.
- Sloan DB, Wu Z, Sharbrough J. 2018. Correction of persistent errors in Arabidopsis reference mitochondrial genomes. Plant Cell. 30(3):525–527. doi: 10.1105/tpc.18.00024.
- Takase M, Zhao T, Zhang M, Chen Y, Liu H, Yang L, Wu X. 2015. An expatiate review of neem, jatropha, rubber and karanja as multipurpose non-edible biodiesel resources and comparison of their fuel, engine and emission properties. Renew Sustain Energy Rev. 43:495–520. doi: 10.1016/j.rser.2014.11.049.
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14(2):178–192. doi: 10.1093/bib/bbs017.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wang Z, Xu B, Li B, Zhou Q, Wang G, Jiang X, Wang C, Xu Z. 2020. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ. 8:e8251. doi: 10.7717/peerj.8251.
- Zhang J, Bai H, Zhang Y. 2020. The complete mitochondrial genome of a mangrove plant: Bruguiera sexangula. Mitochondrial DNA Part B. 5(2):1852–1853. doi: 10.1080/23802359.2020.1750990.
- Zhang J-F, Zhao L, Duan N, Guo H-X, Wang C-Y, Liu B-B. 2019. Complete chloroplast genome of Euphorbia hainanensis (Euphorbiaceae), a rare cliff top boskage endemic to China. Mitochondrial DNA Part B. 4(1):1325–1326. doi: 10.1080/23802359.2019.1596761.
- Zhang S, Bai H, Liu Q, Zhang Y. 2020. The complete mitochondrial genome of a mangrove plant: Bruguiera sexangula (Lour.) Poir. var. rhynchopetala Ko. Mitochondrial DNA Part B. 5(2):1773–1774. doi: 10.1080/23802359.2020.1750316.
- Zhang Y, Shi Y, Duan N, Liu B-B, Mi J. 2019. Complete chloroplast genome of Euphorbia tirucalli (Euphorbiaceae), a potential biofuel plant. Mitochondrial DNA Part B. 4(1):1973–1974. doi: 10.1080/23802359.2019.1617069.
