Abstract
Megalurothrips usitatus (Bagnall, 1913) (Thysanoptera: Thripidae) is a widely distributed pest in Asia that primarily affects the production of snap beans and cowpea. The complete mitochondrial genome of Megalurothrips usitatus has been sequenced and annotated in this study, which is 17,209 bp long and contains 13 protein-coding genes (PCGs), two rRNAs, and 22 tRNA genes. Most of the protein-coding genes (PCGs) start with ATG except ND4 using TTG. Meanwhile, eight PCGs stop with TAA, four PCGs have an incomplete stop codon, and the gene Cytb ends with TAG. Phylogenetic analysis showed that M. usitatus is closely related to Frankliniella intonsa and F. occidentalis, providing a basis for the study of the mitochondrial evolution of Thripinae.
1. Introduction
Megalurothrips usitatus (Bagnall, 1913) (Thysanoptera: Thripidae) is the most significant pest on legumes in Asia (Mound and Kibby Citation1998; Tang et al. Citation2015), causing severe damage to cowpeas and snap beans in southern regions of China. The female M. usitatus is approximately 1.6 mm size and has a brown coloration (as shown in ). It possesses 8-sectioned antennae, and has compound and monocular eyes. The mesosternal furca has a spinula, while the metafurca does not. The narrow wings of the female have fine tassels peripherally. They are colorless near the base 1/4 of the forewing and near the tip, while the middle and tip are brown (Palmer, Citation1987). M. usitatus feeds on young tissues, including growing points, young flowers, and flower buds, causing leaf shrinkage, deformation, and the loss of flowers and pods, ultimately affecting yield (Huang et al. Citation2018; Liu et al. Citation2020). However, identifying this group based on morphological features alone is difficult due to their small size and species abundance, making it necessary to use molecular data accurately for exploring the taxonomic status of each species within this subfamily (Qiu et al. Citation2014). To enhance the comprehension of molecular evolution and taxonomic classification of thrips, we sequenced and characterized the mitochondrial genome of M. usitatus.
2. Materials and methods
2.1. Sample collection
M. usitatus was obtained from Haikou (E110°11′38″, N20°02′45″), Hainan Province, China, in 2020. The specimens were photographed, morphologically identified, stored in absolute ethanol, and placed in a freezer at −20 °C. The specimen was deposited at School of Plant Protection, Hainan University (https://hd.hainanu.edu.cn/zhiwu/xwzx/jx_kyfwpt/bbg.htm, contact person: Jinhua Li; email: [email protected]) under the voucher number Mus-hn-L1.
2.2. Methods
Total genomic DNA was extracted from the 3rd-instar larvae and then sequenced on Illunima NovaSeq 6000 platform. The mitochondrial genome sequence was assembled using NOVAPlasty (2.4 version) and annotated with Geseq (https://chlor-obox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017) and MITOS (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013), and then illustrated by OGDRAW online software (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). To validate the reliability of the genome, a comparison was made with mitogenomes of other Thysanoptera species, and two of them, Aeolothrips xinjiangensis and Franklinothrips vespiformis, were used as outgroups. The complete mitochondrial genome sequences of these 13 insect species were downloaded from NCBI database. Nucleotide sequences of all protein-coding genes and ribosomal genes were individually aligned and concatenated using Geneious Prime 2021 (Kearse et al. Citation2012). MrModeltest version 2.4 (Darriba et al. Citation2020) was used to select the best-fit model. Maximum likelihood phylogenies were inferred using IQTREE (Nguyen et al. Citation2015) under the GTR + I + G model for 1000 bootstraps. Bayesian Inference phylogenies were inferred using MrBayes 3.2.1 (Ronquist et al. Citation2012) based on Markov Chain Monte Carlo (one cold and three hot chains) chains of 2,000,000 with 25% burn-in, and sampling was done every 100 generation. The final average standard deviation of split frequencies was 0.0007. The evolutionary tree is visualized in itol (https://itol.embl.de/) (Letunic and Bork Citation2007).
3. Results
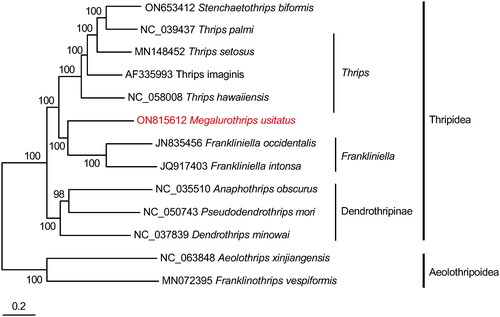
The coverage-depth map was provided, and the average depth was 770 x (Supplementary materials, Figure S1). The complete mitochondrial genome was 17,209 bp long, and the total length of all 13 protein-coding genes (ND1-ND6, ND4L, COX1-COX3, ATP6, ATP8, Cytb), accounting for 62.14% of the whole genome sequence. In addition to these 13 protein-coding genes, the genome contains two rRNAs and 22 tRNA genes (). The tRNA genes are between 56 and 68 bp in length. The total nucleotide composition of the genome is 42.02% A, 11.98% C, 10.20% G, and 35.80% T. The A + T content (77.83%) is significantly higher than that of G + C (22.17%), consistent with most insect mitochondrial genomes (Tyagi et al. Citation2020). Most of the protein-coding genes start with ATN including ATA for ND1, ND5, COX2, Cytb, ATP8, and ATT for ND2, ND3, COX3, ATP6 and ATG for ND4L, ND6, COX1, ATP8, except ND4 which uses TTG. Meanwhile, eight PCGs have TAA as the stop codon, Cytb ends with the unfrequent stop codon TAG, while ND1, ND2, ND4 and ATP8 have an incomplete stop codon T-. Gene editing can be employed to ensure the proper functioning of the stop codon in these cases. The incomplete stop codon T is commonly reported and could produce functional stop codons in polycistronic transcription cleavage and polyadenylation mechanisms (Boore Citation2001). S-rRNA and L-rRNA have a 730 bp and 1162 bp in length for each. Phylogenetic analysis indicated that M. usitatus belongs to the tribe Thripidae, and is closer to its sister clade Frankliniella ().
Figure 2. The mitochondrial genome map of Megalurothrips usitatus. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. GC and AT contents across the mitochondrial genome is shown with dark and light shading, respectively, inside the inner circle.
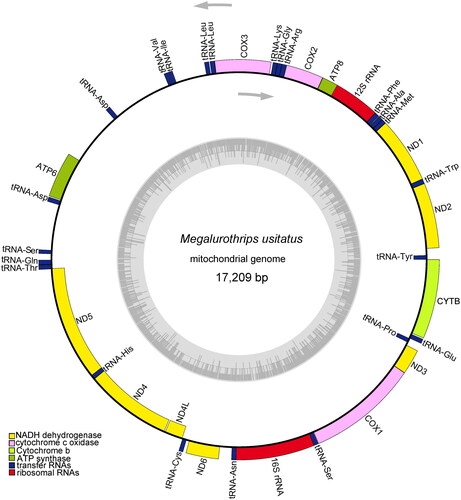
Figure 3. Phylogenetic tree of 13 insect species, including Megalurothrips usitatus based on the nucleotide dataset of the 13 mitochondrial protein-coding genes and ribosomal genes. ‘GTR + I + G’ was used as the best-fit nucleotide substitution model. Bayesian Inference phylogenies were inferred using Markov Chain Monte Carlo (one cold and three hot chains) chains of 2,000,000 with 25% burn-in, and sampling was done every 100 generation. The posterior probabilities are indicated above the nodes. The GenBank accession no. of sequences used in the study are stenchaetothrips biformis ON653412 (Hu et al. Citation2023); Thrips palmi NC_039437 (Chakraborty et al. Citation2018); Thrips setosus MN148452; Thrips imaginis AF335993 (Shao and Barker Citation2003); Thrips hawaiiensis NC_058008; Megalurothrips usitatus ON815612 (this study); Frankliniella occidentalis JN835456 (Yan et al. Citation2012); Frankliniella intonsa JQ917403 (Yan et al. Citation2014); dendrothrips minowai NC_037839; anaphothrips obscurus NC_035510 (Liu et al. Citation2017); pseudodendrothrips mori NC_050743; Aeolothrips xinjiangensis NC_063848; Franklinothrips vespiformis MN072395 (Tyagi et al. Citation2020).
4. Discussion and conclusion
The mitochondrial genome of M. usitatus was 17,209 bp in length, and it contained 13 protein-coding genes (PCGs), 22 tRNAs and 2 rRNA genes. The analysis showed that the genomic structure of M. usitatus is similar to most bilateral animals previously studied (Yan et al. Citation2014). Phylogenetic analysis revealed the phylogenetic position of M. usitatus and indicated that M. usitatus belongs to the tribe Thripidae, a sister clade to the species of Frankliniella. Additionally, our study yields significant insights into the mitogenome of M. usitatus, which will facilitate further research on evolution, germplasm identification, and molecular markers development. Consequently, it is thought that the phylogenetic analysis conducted in this study can distinguish species of Thysanoptera by utilizing mitochondrial gene sequences.
Author contributions
J-H L planned and designed the research. P L collected the insect materials, X-M L performed experiments, D-C C and X-M L analyzed the data. X-M L and P L wrote the manuscript.
Ethics approval and consent to participate
The material used in this study is widely distributed in southern regions of China and does not belong to the IUCN Red List. No specific permits were required for animal collection. The study did not require ethical approval or consent, as no endangered or protected animal species were involved.
Supplemental Material
Download TIFF Image (5 MB)Supplemental Material
Download MS Word (163.8 KB)Disclosure statement
All authors have read and approved the final manuscript. No conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON815612. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA935568, SRP423475 and SAMN33321779, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Boore J-L. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18(7):1413–1416. doi: 10.1093/oxfordjournals.molbev.a003925.
- Chakraborty R, Tyagi K, Kundu S, Rahaman I, Singha D, Chandra K, Patnaik S, Kumar V. 2018. The complete mitochondrial genome of Melon thrips, Thrips palmi (Thripinae): comparative analysis. PLoS One. 13(10):e0199404. doi: 10.1371/journal.pone.0199404.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. doi: 10.1093/molbev/msz189.
- Hu Q-L, Ye Z-X, Zhang C-X. 2023. High-throughput sequencing yields a complete mitochondrial genome of the rice thrips, Stenchaetothrips biformis (Thysanoptera: thripidae). Mitochondrial DNA B Resour. 8(2):204–206. doi: 10.1080/23802359.2023.2169572.
- Huang W-K, Kong X-Y, Ke Y-C, Wang S, Li Q-J, Fu Q-W, Wu Q-X, Liu Y. 2018. Research progress on Thrips Megalurothrips usitatus. China Vegetables. 348(2):21–27.doi: 10.19928/j.cnki.1000-6346.2018.02.005.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Letunic I & Bork P. 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128. doi: 10.1093/bioinformatics/btl529.
- Liu H-R, Li H, Song F, Gu W-Y, Feng J-N, Cai W-Z, Shao R-F. 2017. Novel insights into mitochondrial gene rearrangement in thrips (Insecta: thysanoptera) from the grass thrips Anaphothrips obscurus. Sci Rep. 7(1):4284. doi: 10.1038/s41598-017-04617-5.
- Liu P-P, Qin Z-F, Feng M-Y, Zhang L, Huang X-Z, Shi W-P. 2020. The male-produced aggregation pheromone of the bean flower thrips Megalurothrips usitatus in China: identification and attraction of conspecifics in the laboratory and field. Pest Manag Sci. 76(9):2986–2993. doi: 10.1002/ps.5844.
- Mound LA, Kibby G. 1998. Thysanoptera: An identification guide. Oxford and New York: Ed. CABI p. 70.
- Nguyen L-T, Schmidt H-A, Von Haeseler A, Minh B-Q. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. doi: 10.1093/molbev/msu300.
- Palmer J-M. 1987. Megalurothrips in the flowers of tropical legumes: a morphometric study. In Holman J, Pelikan J, Dixon AFG & Weismann L. [eds] Population structure, genetics and taxonomy of aphids and Thysanoptera. The Hague (SPB Academic Publishing): p. 480–495.
- Qiu H-Y, Liu K, Li P, Fu B-L, Tang L, Zhang M. 2014. Biological characteristics of the bean flower thrips, Megalurothrips usitatus (Thysanoptera: thripidae). Chinese J Trop Crops. 35(12):2437–2441.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Shao R, Barker SC. 2003. The highly rearranged mitochondrial genome of the plague thrips Thrips imaginis (Insecta: thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 20(3):362–370. doi: 10.1093/molbev/msg045.
- Tang L-D, Yan K-L, Fu B-L, Wu J-H, Liu K, Lu Y-Y. 2015. The life table parameters of Megalurothrips usitatus (Thysanoptera: thripidae) on four leguminous crops. Fla Entomol. 98(2):620–625. doi: 10.1653/024.098.0235.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E-S, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic acids research. 45(1): 6–11. doi: 10.1093/nar/gkx391.
- Tyagi K, Chakraborty R, Cameron SL, Sweet AD, Chandra K, Kumar V. 2020. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci Rep. 10(1):695. doi: 10.1038/s41598-020-57705-4.
- Yan D-K, Tang Y-X, Hu M, Liu F-Q, Zhang D-F, Fan J-Q. 2014. The mitochondrial genome of Frankliniella intonsa: insights into the evolution of mitochondrial genomes at lower taxonomic levels in Thysanoptera. Genomics. 104(4):306–312. doi: 10.1016/j.ygeno.2014.08.003.
- Yan D-K, Tang Y-X, Xue X-F, Wang M-H, Liu F-Q, Fan J-Q. 2012. The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: thripidae) contains triplicate putative control regions. Gene. 506(1):117–124. doi: 10.1016/j.gene.2012.06.022.

