Abstract
Allium wallichii Kunth is a herb species with potentially extensive applications because of its edible, ornamental, and pharmaceutical values. The structural characteristics and phylogenetic relationships of its chloroplast genome were determined here for the first time. The complete cp genome was found to be 152,496 bp long, with a GC content of 37.04%. It consists of four distinct regions: a large single copy (LSC) region of 82,510 bp, a small single copy (SSC) region of 17,460 bp, and two inverted repeat (IR) regions of 26,263 bp each. The genome encodes 129 genes, including 86 protein-coding genes, 37 tRNA genes, and six rRNA genes. Our phylogenetic analysis revealed that A. wallichii was closely related to Allium wallichii var. platyphyllum, which are included in the section Bromatorrhiza, subgenus Amerallium Traub of the genus Allium. Our report provides valuable information on the genetic diversity of Allium species.
Introduction
Allium wallichii Kunth (Allium wallichii Kunth Enum. Pl. 1843) is an upright, bulbous, perennial herb belonging to the family Amaryllidaceae. It rises a triangular, columnar, naked scape that bears loose umbels of star-shaped, purple, pink, or white florets in late summer and early autumn from the center of each basal rosette (). It is native to East Asia, West China, and the Himalayas, and is widely distributed on grassy slopes or ditch sides at an elevation of 2300–4800 meters (m) in temperate regions such as Yunnan, Guangxi, Sichuan, and Guizhou Provinces (Jiang and Ge Citation2014). The largest known habitat of wild A. wallichii is located in Jiucaiping planting areas, which are the highest point in Guizhou Province, at an elevation of 2900 m (Chen et al. Citation2021). The natural ecological environment of A. wallichii on Jiucaiping Mountain is shown in . A. wallichii with purple inflorescences is the most common on Jiucaiping Mountain ().
Figure 1. (A) Natural ecological environment in the Allium wallichii Kunth planting area. (B) Morphological characteristics of Allium wallichii Kunth inflorescences. The flowerhead of Allium wallichii Kunth produces a globe-shaped cluster of small, star-shaped flowers that are typically pink or purplish in color. The leaves of that are green, relatively narrow, typically strap-like and arise from the base of the plant. Species photos were taken by the author in Jiucaiping Mountain, Hezhang, Guizhou province, without any copyright issues.

A. wallichii has rich secondary metabolite profiles that have proven medicinal applications because of the effect of dispelling wind and activating blood circulation (Zhao et al. Citation2015). Moreover, it is considered a plant resource of high ornamental value owing to its large tidbits and beautiful and diverse flower colors.
To our knowledge, the chloroplast (cp) genome of A. wallichii has not yet been studied. Therefore, in the present study, the complete cp genome of A. wallichii was sequenced, assembled, and annotated to understand the genetic diversity of Allium species.
Materials and methods
A sample of A. wallichii was legally collected from the Jiucaiping Mountain in Hezhang, Guizhou, China (104°77′34.4′′E, 26°98′79.34′′N). A specimen was deposited at Guizhou University under the voucher number GZU20220701 (contact person: Jin Zhao; email: [email protected]). Total genomic DNA was isolated from the mature leaves using a proprietary DNA-seq kit on the Illumina platform following the manufacturer’s instructions. The total genome sequence was generated by PE150 sequencing on an Illumina HiSeq 2500 platform with insert sizes of 350 bp (Illumina, San Diego, CA, USA). The complete cp genome was assembled using GetOrganelle (Jin et al. Citation2020). Sequence annotation was performed using CPGAVAS2, with Allium hookeri (MZ557488.1) as a reference (Shi et al. Citation2019).
To validate the phylogenetic position of A. wallichii, a phylogenetic tree was constructed based on the complete cp genomes of 62 species, with Narcissus poeticus (NC_039825) as the outgroup. The sequences were aligned using MAFFT-7.745 (Katoh and Standley Citation2013), and jModelTest-2.1.10 (David Citation2008) was used to analyze the best nucleotide substitution model. A maximum likelihood (ML) tree was constructed using RAxML-8.2.12 (Stamatakis Citation2014) with a bootstrap of 1000 replicates.
Results
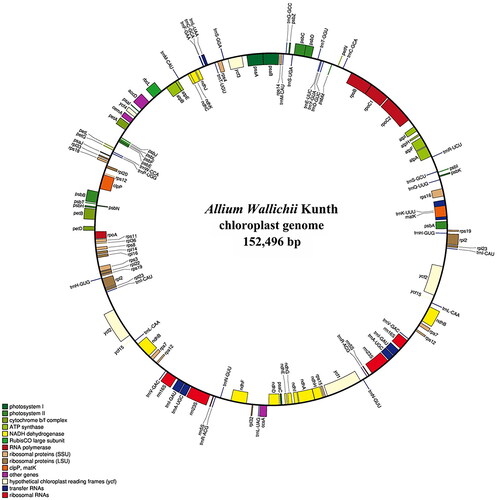
Complete cp genomes were assembled using GetOrganelle (Figure S1). The complete cp genome of A. wallichii (GenBank accession No. ON872149.1) was 152,496 bp long. It consists of four distinct regions: a large single copy (LSC) region of 82,510 bp, a small single copy (SSC) region of 17,460 bp, and two inverted repeat (IR) regions of 26,263 bp. The genome comprised 129 genes, including 86 protein-coding genes, 37 tRNA genes, and six rRNA genes. The overall GC content of the A. wallichii cp genome was 37.04%, while the corresponding values of the LSC, SSC, and IR regions were 34.86%, 30.32%, and 42.71%, respectively. The complete cp map of A. wallichii is shown in .
Figure 2. Gene map of Allium wallichii Kunth chloroplast genome. Genes on the circle are transcribed in the clockwise direction. Genes belonging to different functional groups are indicated by different colors.
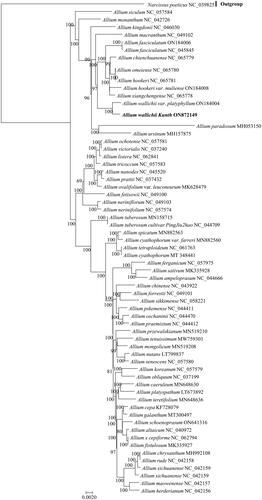
To gain insight into the phylogenetic position of the studied species, a phylogenetic analysis was performed using the complete cp genomes of A. wallichii and 61 other Amaryllidaceae species (). The sequences were aligned using MAFFT and a phylogenetic tree was constructed using RAxML. For the maximum likelihood (ML) analysis, the best-fit model GTRGAMMAI was used, as suggested, with 1000 bootstrap replicates. The results indicated that A. wallichii is closely related to A. wallichii var. platyphyllum. In addition, A. wallichii was included in the section Bromatorrhiza, which belongs to the subgenus Amerallium Traub of the genus Allium. These findings provide a foundation for further studies on the evolutionary relationships of A. wallichii.
Figure 3. Maximum likelihood phylogenetic tree based on the complete chloroplast genomes of 62 species of Amaryllidaceae, with Narcissus poeticus as the outgroup. The tree was constructed by RAxML using the GTRGAMMAI substitution model. Numbers on the nodes are bootstrap values from 1000 replicates. All sequences were downloaded from NCBI’s GenBank.
Discussion
Analysis of the cp genome has been used to investigate the genetic diversity of plant species. However, knowledge on the Allium cp genome is scarce, especially regarding A. wallichii. Thus, in the present study, we assembled its complete cp genome and constructed a phylogenetic tree to systematically analyze the genetic relationship of A. wallichii germplasms with other Amaryllidaceae species. Our phylogenetic analysis showed that A. wallichii formed a sister relationship with A. wallichii var. platyphyllum. A. wallichii, along with 13 Allium species, such as A. fasciculatum, A. hookeri, and A. chienchuanense, was clustered into a monophyletic clade with strong support. The investigated A. wallichii was included in the section Bromatorrhiza, which belongs to the subgenus Amerallium of the genus Allium. The Bromatorrhiza species belonging to this section mainly inhabit wet grassy slopes, areas near rocks, or forest edges, and are widely distributed in the Himalayas–Hengduan mountain region and Qinghai–Tibetan Plateau in China. A. wallichii and several closely related species belonging to the section Bromatorrhiza are alpine species. It has been predicted that this is related to the ecological evolution and adaptation of alpine plants.
Conlusion
In summary, in the present study, we reported the complete cp genome sequence of A. wallichii for the first time, and we analyzed its phylogenetic relationships with other species in the family Amaryllidaceae. Our results provide a reference for genetic evaluation of A. wallichii.
Ethical approval
This study does not address any ethical issues. The collection of plant samples was legal and reasonable.
Geolocation information
Samples were obtained from Jiucaiping Mountain in Guizhou Province, China (26° 59′ 42″ N, 104° 45′ 36″ E). A specimen was deposited at Guizhou University under voucher number GZU20220701 (contact person: Jin Zhao; email: [email protected]).
Author contributions
Jin Zhao conceived the study and wrote and revised the manuscript critically. Miao Huang and Rui Ni wrote a part of the first draft and collected sequence information for the phylogenetic tree. Xiaoyulong Chen and Ting Peng collected the plant materials and revised the manuscript for intellectual content. Hong Nan analyzed the data and performed the chloroplast genome assembly and annotation. All authors agree to be accountable for all aspects of this study.
Supplemental Material
Download JPEG Image (24.4 KB)Supplemental Material
Download PDF (317.9 KB)Acknowledgements
We thank the Institute of Pepper Industry and Technology for supporting this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/nuccore/ON872149) under accession No. ON872149. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA864028, SRR20722597, and SAMN30071150, respectively.
Additional information
Funding
References
- Chen X, Krug L, Yang M, Berg G, Cernava T. 2021. The Himalayan onion (Allium wallichii Kunth) harbors unique spatially organized bacterial communities. Microb Ecol. 82(4):909–918. doi:10.1007/s00248-021-01728-5.
- David P. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Jiang K, Ge D. 2014. Preliminary report on plant resources and community ecological characteristics of chives in Hezhang county. Ningxia Agric For Sci Technol. 55(10):38–39.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Zhao C, Gong X, Wang D, et al. 2015. Chemical constituents of volatile oil by SPME from different parts of Allium wallichii. Food Sci. Tech. 40(4):325–328.
