Abstract
Ocimum americanum Linnaeus 1755 (Lemon basil) is an essential medicinal species in the Ocimum genus. Its leaf decoction is traditionally used to treat diabetes, constipation, diarrhea, piles, and dysentery. The essential oils from this species have intense fungicidal activity. The complete chloroplast genome sequence of O. americanum was assembled from Illumina paired-end sequencing data. The O. americanum chloroplast genome was 152,460 bp in length, containing a large single copy (LSC) region of 83,459 bp and a small single copy (SSC) region of 17,607 bp, separated by a pair of inverted repeats (IRs) of 25,697 bp. The genome contained 134 unique genes, including 89 protein-coding, 37 tRNA, and eight rRNA genes. Among them, nine genes had a single intron, and two genes contained two introns. The overall GC content of the chloroplast genome was 38%, while the corresponding values of LSC, SSC, and IR regions were 35.8%, 31.7%, and 43.1%, respectively. In the phylogenetic analysis, all the Ocimum species formed a group closely related to Plectranthus barbatus. O. americanum was more closely related to O. gratissimum and O. basilicum than the other species of Ocimum included in this study.
Introduction
Ocimum americanum Linnaeus 1755 (Lemon Basil) is a potential traditional medicinal plant species. The decoction from the leaves of this species is commonly used for treating diabetes, constipation, diarrhea, piles, and dysentery (Bassole et al. Citation2005). The pharmacological activities of O. americanum are antioxidant, antifungal, antimicrobial, insecticidal, larvicidal, and gastric cytoprotective (Javanmardi et al. Citation2003, Vidhya et al. Citation2020). The estimated consumption of O. americanum as herbal medicine was 500–1000 MT (in dried condition). It is possibly used as a substitute or adulterant instead of Ocimum tenuiflorum (Ravikumar et al. Citation2018). The genus Ocimum contains around 65 species, and the key species used in herbal medicine are O. americanum, O. basilicum, O. tenuiflorum, and O. gratissimum. Classification of Ocimum species is challenging due to their significant morphological similarities and lack of genetic diversity. Species relationships were studied using morphological characters (Sobti and Pushpangadan Citation1979), pollen morphology (Harley et al. Citation1992), and DNA markers (Singh et al. Citation2004, Chen et al. Citation2013, Kumar et al. Citation2016). Singh et al. (Citation2004) used Random Amplified Polymorphic DNA (RAPD) markers and classified five Ocimum species into two groups: one with O. tenuiflorum and O. gratissimum, and the other with O. basilicum, O. kilimandscharicum, and O. americanum. However, Chen et al. (Citation2013) reported that O. gratissimum was closely related to O. americanum and O. basilicum. Based on the analysis of the psbA-trnH sequence, Kumar et al. (Citation2016) classified the Ocimum species into Gratissimum group with O. gratissimum and O. americanum and Sanctum group with O. tenuiflorum and O. adscendens. In this study, we sequenced and assembled the complete chloroplast genome of O. americanum using Illumina paired-end sequencing data to contribute to the systematics, DNA barcoding, molecular markers, and phylogenetic analysis within the Ocimum genus.
Materials and methods
Fresh plant material of O. americanum () was collected from the wasteland in Ozhavetti village, Madhurandhagam, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°29′49.4″N 79°52′32.9″E). The voucher specimen (MH178052) was deposited in the Madras Herbarium, Botanical Survey of India (BSI), Southern Regional Centre, Coimbatore, Tamil Nadu, India (https://bsi.gov.in/regional-centres/en?rcu=133, Dr. M. U. Sharief, Scientist-E and Head of Office, email: [email protected]). O. americanum is a small and erect plant with lanceolate or ovate-lanceolate leaves, Apex of the leaf is acute, the margin is entire or sparsely serrate, and leaves are pubescent beneath with long hairs on midrib and lateral veins. Stems are quadrangular in shape and purple to brown in color, and the flowers are white (Suddee et al. Citation2004). Total genomic DNA from O. americanum was extracted following the Cetyltrimethyl ammonium bromide method (Doyle and Doyle Citation1987), with minor modifications (Nithaniyal et al. Citation2014). According to the manufacturer’s procedure, a paired-end DNA library was constructed using the Nextera XT Library Prep Kit (Cat. No. FC-131-1024). The library was sequenced on the Illumina Novoseq 6000 platform (Illumina Inc., San Diego, CA) with a paired-end sequencing length of 150 bp. Paired-end sequencing generated 2.3 Gb data with q30 bases of more than 87%. The chloroplast genome of O. americanum was assembled using NovoPlasty (k-mer 31) with O. gratissimum L. (Balaji et al. Citation2021) as a reference seed sequence (Dierckxsens et al. Citation2017). The assembled complete chloroplast genome of O. americanum was annotated with GeSeq (Tillich et al. Citation2017). The predicted transfer RNAs (tRNAs) were identified by tRNAscan-SE 2.0 (Lowe and Chan Citation2016). In addition, the CPGView (www.1kmpg.cn/cpgview/; Liu et al. Citation2023) was applied to structures to visualize the intron-containing genes. The sequencing depth of the assembled chloroplast genome was done by aligning to the raw reads using a BWA aligner (Li and Durbin Citation2009). The bam file was viewed using the software Qualimap for the coverage map (Fernando et al. Citation2012). A phylogenetic tree was constructed with 1000 bootstrap replicates using RAxML (Random Axelerated Maximum Likelihood) version 8 (Stamatakis Citation2014) from the alignments created using the MAFFT program (Katoh and Standley Citation2013).
Results and discussion
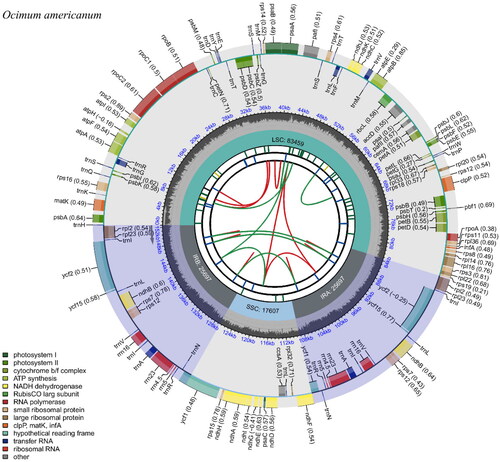
The assembled complete chloroplast genome sequence of O. americanum was 152,460 bp with a mean coverage of 128×. The circular map of the chloroplast genome is shown in . It showed a typical quadripartite structure, including an LSC (large single copy) region of 83,459 bp, an SSC (small single copy) region of 17,607 bp, and a pair of inverted repeats (IRs) of 25,697 bp. The overall GC content was 38%, while LSC, SSC, and IR regions were 35.8%, 31.7%, and 43.1%, respectively. The chloroplast genome contained 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Nine genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16) were with a single intron, and two genes (pafI and clpP) contained two introns (Supplementary Figures 1 and 2). The minimum, maximum, and average depth of coverage of the assembled chloroplast genome was 10×, 697×, and 109×, respectively (Supplementary Figure 3). The complete chloroplast genome of O. americanum with annotations was submitted to GenBank (Accession No. OQ689804). The raw reads were deposited to NCBI under the Sequence Read Archive database (Accession No. SRR23955912).
Figure 2. Circular map of the chloroplast genome of Ocimum americanum. From the center going outward, the first circle shows the distribution of the repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle displays the tandem repeats marked with short bars. The third circle shows the LSC, SSC, IRa, and IRb regions. The fourth circle shows the percentage of GC content. The next circle shows the genes having different colors based on the functional groups. The functional classification is shown at the bottom left. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counter-clockwise direction.
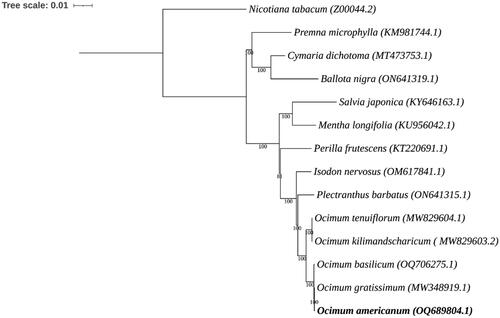
The complete chloroplast genome sequences of 13 species were retrieved from the National Center for Biotechnology Information (NCBI) to conduct the phylogenetic analysis. The complete chloroplast genome sequences were subjected to multiple sequence alignment using the MAFFT tool. The phylogenetic tree was constructed using RAxML (Random Axelerated Maximum Likelihood; Stamatakis Citation2014). The tree included the chloroplast genomes of 14 species, including O. americanum from this study, 13 from the Lamiaceae family, and N. tabacum from Solanaceae as an outgroup. The complete chloroplast genome sequences were utilized to understand the phylogenetic relationships among species (). All the species of Ocimum formed a clade, and this group was closely related to Plectranthus barbatus. O. americanum was closely related to O. gratissimum and O. basilicum within Ocimum. This relationship was consistent with a previous report based on RAPD and ISSR markers (Chen et al. Citation2013). The application of chloroplast genome-based analysis demonstrated a more robust species resolution within the Lamiaceae family compared to the resolutions observed using morphological characters (Sobti and Pushpangadan Citation1979) and RAPD markers (Singh et al. Citation2004). This complete chloroplast genome of O. americanum can be subsequently used for phylogenetic analysis, DNA barcoding, and authentication of lemon basil.
Figure 3. Phylogenetic tree based on the whole chloroplast genome sequences of 13 species from Lamiaceae family, including the chloroplast genome of O. americanum (OQ689804) from this study. Nicotiana tabacum (Solanaceae) was used as an outgroup. The bootstrap support values are shown on the nodes. The following sequences were used: O. americanum OQ689804.1 (this study), O. gratissimum MW348919.1 (Balaji et al. Citation2021), O. basilicum OQ706275.1 (Rabah et al. Citation2017), O. tenuiflorum MW829604.1 (Harini et al. Citation2021), O. kilimandscharicum MW829603.2 (Renald et al. Citation2021), Plectranthus barbatus ON641315.1, Isodon nervosus OM617841.1, Perilla frutescens KT220691.1, Mentha longifolia KU956042.1 (Vining et al. Citation2017), Salvia japonica KY646163.1 (He et al. Citation2017), Premna microphylla KM981744.1 (Yang and Kong Citation2016), Ballota nigra ON641319.1, Cymaria dichotoma MT473753.1 (Zhao et al. Citation2021), and Nicotiana tabacum Z00044.2 (Shinozaki et al. Citation1986).
Conclusion
In this study, the chloroplast genome sequence of Ocimum americanum was assembled and annotated for the first time. The phylogenetic analysis showed that the chloroplast genome of O. americanum is closely related to O. gratissimum and O. basilicum within the Lamiaceae family. This study provides valuable chloroplast genome resources of the genus Ocimum, which lay the foundation for the study of phylogenetic relationships within the Lamiaceae family and the development of species-specific molecular markers for plant authentication of herbal drugs.
Author contributions
S. Vineesh, R. Balaji, and Tanuja collected the specimen material, conducted the experiment, analyzed the sequence data, and drafted the paper. M. Parani contributed to the conception and design of this work. All the authors carefully read, revised, and approved the final manuscript to be published. We thank Dr. D. Narasimhan for his help in the authentication of plant material and for providing comments and suggestions on the final manuscript.
Ethical approval
No permissions were required for the sample collection of Ocimum americanum L. because it is widely distributed in the wastelands and roadsides in tropical regions. The plant species was collected from Ozhavetti village, Madhurandhagam, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°29′49.4″N 79°52′32.9″E).
Supplemental Material
Download TIFF Image (74.2 MB)Supplemental Material
Download TIFF Image (3.7 MB)Supplemental Material
Download TIFF Image (173.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data supporting this study’s findings are publicly available in the NCBI database. The complete chloroplast genome of O. americanum was deposited in GenBank under the accession number OQ689804 (https://www.ncbi.nlm.nih.gov/nuccore/OQ689804.1). The next-generation sequencing data files are available from the SRA, Bio-Sample, and BioProject ID under the accession numbers PRJNA948343, SAMN33896533, and SRR23955912, respectively.
Additional information
Funding
References
- Balaji R, Ravichandiran K, Tanuja , Parani M. 2021. The complete chloroplast genome of Ocimum gratissimum from India – a medicinal plant in the Lamiaceae.Mitochondrial DNA B Resour. 6(3):948–950. doi: 10.1080/23802359.2021.1889413.
- Bassole IHN, Nebie R, Savadogo A, Ouattara CT, Barro N, Traore SA. 2005. Composition and antimicrobial activities of the leaf and flower essential oils of Lippia chevalieri and Ocimum canum from Burkina Faso. Afr J Biotechnol. 4(10):1156–1160.
- Chen SY, Dai TX, Chang YT, Wang SS, Ou SL, Chuang WL, Chuang CY, Lin YH, Lin YH, Ku HM. 2013. Genetic diversity among Ocimum species based on ISSR, RAPD and SRAP markers. Aust J Crop Sci. 7(10):1463–1471.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fernando G, Konstantin O, José C, Luis MC, Stefan G, Sonia T, Joaquín D, Thomas FM, Ana C. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 28(20):2678–2679.
- Harini P, Balaji R, Parani M. 2021. The complete chloroplast genome of Ocimum tenuiflorum L. subtype Rama Tulsi and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(8):2224–2226. doi: 10.1080/23802359.2021.1944381.
- Harley MM, Paton A, Harley RM, Cade PG. 1992. Pollen morphological studies in tribe Ocimeae (Nepetoideae: labiatae): I. Ocimum L. Grana. 31(3):161–176. doi: 10.1080/00173139209432027.
- He YH, Han LM, Liu YP, Tian N, Su X, Wang ZZ. 2017. Complete sequence analysis of chloroplast genome of Salvia japonica. Bull Bot Res. 37(4):572–578.
- Javanmardi J, Stushnoff C, Locke E, Vivanco JM. 2003. Antioxidant activity and total phenolic content of Iranian Ocimum accessions. Food Chem. 83(4):547–550. doi: 10.1016/S0308-8146(03)00151-1.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kumar A, Mishra P, Baskaran K, Shukla AK, Shasany AK, Sundaresan V. 2016. Higher efficiency of ISSR markers over plastid psbA-trnH region in resolving taxonomical status of genus Ocimum L. Ecol Evol. 6(21):7671–7682. doi: 10.1002/ece3.2483.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi: 10.1093/nar/gkw413.
- Nithaniyal S, Newmaster SG, Ragupathy S, Krishnamoorthy D, Vassou SL, Parani M. 2014. DNA barcode authentication of wood samples of threatened and commercial timber trees within the Tropical Dry Evergreen Forest of India. PLoS One. 9(9):e107669. doi: 10.1371/journal.pone.0107669.
- Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JSM, Jansen RK, Ruhlman TA. 2017. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome. 10(3):1–14. doi: 10.3835/plantgenome2017.03.0020.
- Ravikumar K, Begum SN, Ved DK, Bhatt JR, Goraya GS. 2018. Compendium of traded medicinal plants. Bengaluru (India): Foundation for Revitalization of Local Health Traditions (FRLHT); p. 104.
- Renald YS, Balaji R, Tanuja , Parani M. 2021. The complete chloroplast genome and phylogenetic analysis of Ocimum kilimandscharicum Gurke (Camphor Basil) from India. Mitochondrial DNA B Resour. 6(8):2164–2165.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x.
- Singh AP, Dwivedi S, Bharti S, Srivastava A, Singh V, Khanuja SPS. 2004. Phylogenetic relationships as in Ocimum revealed by RAPD markers. Euphytica. 136(1):11–20. doi: 10.1023/B:EUPH.0000019497.89760.e8.
- Sobti SN, Pushpangadan P. 1979. Cytotaxonomical studies in the genus Ocimum. In: Bir SS, editor. Taxonomy, cytogenetics and cytotaxonomy of plants. New Delhi (India): Kalyani Publishers; p. 373–377.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Suddee S, Paton AJ, Parnell JAN. 2004. Taxonomic revision of tribe Ocimeae Dumort. (Lamiaceae) in Continental South East Asia III. Ociminae. Kew Bulletin. 59(3):379. doi: 10.2307/4110950.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Vidhya E, Vijayakumar S, Rajalakshmi S, Kalaiselvi S, Pandiyan P. 2020. Antimicrobial activity and phytochemical screening of Ocimum americanum L. extracts against pathogenic microorganisms. Acta Ecologica Sinica. 40(3):214–220. doi: 10.1016/j.chnaes.2019.09.001.
- Vining KJ, Johnson SR, Ahkami A, Lange I, Parrish AN, Trapp SC, Croteau RB, Straub SCK, Pandelova I, Lange BM. 2017. Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Mol Plant. 10(2):323–339. doi: 10.1016/j.molp.2016.10.018.
- Yang J, Kong W. 2016. The complete chloroplast genome sequence of Premna microphylla Turcz. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4164–4165. doi: 10.3109/19401736.2014.1003894.
- Zhao F, Chen YP, Salmaki Y, Drew BT, Wilson TC, Scheen AC, Celep F, Bräuchler C, Bendiksby M, Wang Q, et al. 2021. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 19(1):2. doi: 10.1186/s12915-020-00931-z.

