Abstract
The complete mitochondrial genome of Polythlipta liquidalis Leech, 1889 was sequenced and annotated in this study, which was the first reported complete mitogenome of the genus Polythlipta. The mitogenome of P. liquidalis is 15,305 bp in length and was predicted to encode 37 typical mitochondrial genes including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and one major non-coding A-T rich region. The maximum likelihood phylogenetic analysis based on the 13 PCGs was constructed, including P. liquidalis and 15 related Spilomelinae species, using Ostrinia furnacalis as the outgroup. The result showed that P. liquidalis is grouped with Sinomphisa plagialis. These data will serve as a molecular resource for species identification of P. liquidalis and become a valuable resource for a range of genetic, functional, evolutionary and comparative genomic studies on members of Spilomelinae.
1. Introduction
Polythlipta liquidalis Leech, 1889 is widely distributed in China, Japan and North Korea. This species is subordinate to the subfamily Spilomelinae of Crambidae, which contains a variety of species that can bring about great economic loss to agriculture and forestry. The genetics structure and phylogenetic status of the genus Polythlipta have not been investigated before, therefore the complete mitochondrial genome of P. liquidalis was characterized for the first time in this study, which would provide useful genomic data for future phylogenetic and taxonomic classification of the Spilomelinae.
2. Materials and methods
The specimen of P. liquidalis was collected by light trap in July 2018 from Mount Emei (29.59°N, 103.38°E), Sichuan Province, China. The specimen was deposited at the Entomological Museum in the College of Bioscience and Engineering, Jiangxi Agricultural University under the voucher number MT001 (https://www.jxau.edu.cn/, Hua Rong, [email protected]).
The species P. liquidalis can be well diagnosed by the remarkable wing pattern as follows (): Both the forewing and hindwing are white. The basal 1/3 of the forewing has a yellow-brown triangle patch which is surrounded by fuscous line; a large black patch is located at upper 2/3 of the distal part. The hind wing has a blackish brown ribbonlike discocellular spot (Li et al. Citation2012).
Figure 1. External features of P. liquidalis. This photo was taken by Hua Rong with the author’s approval for use.

Total genomic DNA was extracted from thoracic muscle using the HiPure Insect DNA Kit (Magen, China). Libraries with an average fragment length of 350 bp were constructed. Then, the whole-genome sequencing was performed using the Illumina NovaSeq 6000 platform, and approximately 6 Gb raw data was obtained. After data quality control, the clean data was assembled into the complete mitogenome and annotated employing MitoZ v2.4-alpha (Meng et al. Citation2019). Furthermore, manual correction was performed to ensure annotation accuracy using MITOS2 (Donath et al. Citation2019). Finally, the mitochondrial genome cycle map of P. liquidalis was drawn with the program proksee v.6.0.2 (https://proksee.ca/) (Grant et al. Citation2023).
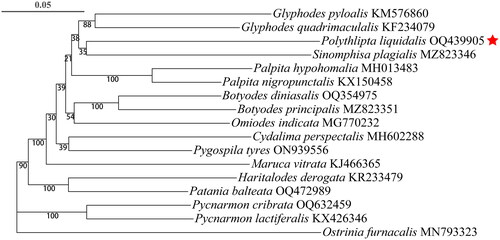
To analyze the phylogenetic position of P. liquidalis, the complete sequences of mitochondrial genomes of the fifteen closest species were downloaded based on the blasting results in GenBank, and Ostrinia furnacalis was selected as the outgroup (). Nucleotide sequences for each of the 13 PCGs were translated into amino acids, aligned separately with MAFFT implemented in PhyloSuite v1.2.2, and then toggled back into nucleotide alignments. The individual nucleotide alignments were concatenated using PhyloSuite v1.2.2. Based on the concatenated alignment, the phylogenetic tree was generated using the maximum likelihood (ML) analyses with 1000 bootstrap replicates in PhyloSuite v1.2.2 (Zhang et al. Citation2020).
Table 1. Species and GenBank accession number of mitogenomes used in this study.
3. Results
The complete mitochondrial genome sequence of P. liquidalis was deposited in GenBank under accession number OQ439905. It was a closed circular molecular structure of 15,305 bp in length, with an average depth of 605.2× (Supplementary Figure S1). For the low coverage depth of the AT-rich region around 15,000 bp, Sanger sequencing was further performed, and the result was consistent with that of NGS assembly, indicating the correctness of this mitochondrial genome assembly data (Supplementary Figure S2). It contains 13 protein coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12S rRNA and 16S rRNA), and a non-coding control region (A-T rich region) (). The order and orientation of the mitochondrial genes are identical to the inferred ancestral arrangement of Lepidoptera. The nucleotide composition is A 40.3%, C 11.5%, G 7.5%, and T 40.7%, showing a highly A/T bias as commonly present in insects (Boore Citation1999).
Figure 2. Mitochondrial genome map of P. liquidalis. Represented with arrows, the transcription directions for the outer and inner genes are listed clockwise and anticlockwise, respectively.
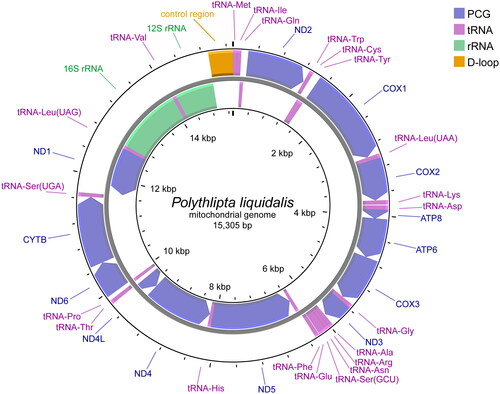
The total length of the 13 PCGs is 11,199 bp, accounting for 73.2% of the whole genome. Most PCGs initiate with typical mitochondrial start codon ATN except for COX1 with TTG, as four PCGs (ND2, ND3, ND5 and ND4L) with ATT, six (COX2, ATP6, COX3, ND4, CYTB and ND1) with ATG, and two (ATP8 and ND6) with ATC. Ten of the 13 PCGs terminated with complete stop codon TAA, whereas COX1 and COX2 used single T nucleotide, and ND5 used TA nucleotides as an incomplete stop codon. The lengths of 22 tRNAs ranged from 63 bp (tRNA-Arg) to 71 bp (tRNA-Lys and tRNA-Asp). Two rRNA genes (12S rRNA and 16S rRNA), which are divided by tRNA-Val, are 1348 bp and 782 bp, respectively.
The ML phylogenetic tree showed that the P. liquidalis was firstly clustered with Sinomphisa plagialis, then together with Glyphodes pyloalis and Glyphodes quadrimaculalis ().
4. Discussion and conclusion
By this work, the complete mitogenome of P. liquidalis was identified for the first time. It was similar to that of other Lepidoptera members in relation to gene organization and composition (Liu et al. Citation2021). In the ML phylogenetic tree, P. liquidalis is grouped with S. plagialis. These results will be helpful for understanding the systematics among members of the subfamily Spilomelinae.
We noticed that the read counts were extremely low in the high AT-content control region of the mitogenome. This phenomenon has also been reported in other studies (Oyola et al. Citation2012; Gan et al. Citation2019), suggesting a bias of Illumina sequencing toward high AT-content regions. One possible explanation may be that the DNA fragment with high AT-content inhibited the amplification of DNA polymerase in sequencing reactions. For sequencing of the AT-rich region, it might be a feasible strategy to improve the representation of extremely high AT-content DNA fragments by lowering the annealing and extension temperature during library preparation. Alternatively, a PCR-free library preparation is also an option to obtain uniform coverage depth of genome with high AT-content (Williams et al. Citation2012). We hope this finding will be instructive for the subsequent Illumina sequencing and assembly of mitochondrial genomes.
Ethical approval
The permission for collecting the sample was received from the Scenic Area Management Committee of Mount Emei. And the experiments were performed in accordance with the recommendations of the Ethics Committee for Animal Experiments of Jiangxi Agricultural University. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
HR and YW collected the samples; HR and LDY conceived and designed the experiments; LDY, RHW and YYZ performed the experiments; YW analyzed and interpreted the data; LDY wrote the manuscript; YW and HR revised the manuscript. All authors read, revised, and approved the final manuscript and agreed to be accountable for the work.
Supplemental Material
Download MS Word (566.5 KB)Acknowledgments
The authors thank Chen Yang for the bioinformatics support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OQ439905. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA935109, SRR23461129, and SAMN33298022, respectively.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. doi:10.1093/nar/27.8.1767.
- Chen S, Li FH, Lan XE, You P. 2016. The complete mitochondrial genome of Pycnarmon lactiferalis (Lepidoptera: crambidae). Mitochondrial DNA B Resour. 1(1):638–639. doi:10.1080/23802359.2016.1214551.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Martin M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi:10.1093/nar/gkz833.
- Gan HM, Linton SM, Austin CM. 2019. Two reads to rule them all: nanopore long read-guided assembly of the iconic Christmas Island red crab, Gecarcoidea natalis (Pocock, 1888), mitochondrial genome and the challenges of AT-rich mitogenomes. Mar Genomics. 45:64–71. doi:10.1016/j.margen.2019.02.002.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi:10.1093/nar/gkad326.
- Li HH, et al. 2012. Microlepidoptera of Qinling Mountains. Insecta: lepidoptera. Beijing: science Press.
- Liu XM, Qi MJ, Xu HZ, Wu ZP, Hu LZ, Yang MS, Li HH. 2021. Nine Mitochondrial Genomes of the Pyraloidea and Their Phylogenetic Implications (Lepidoptera). Insects. 12(11):1039. doi:10.3390/insects12111039.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63. doi:10.1093/nar/gkz173.
- Oyola SO, Otto TD, Gu Y, Maslen G, Manske M, Campino S, Turner DJ, MacInnis B, Kwiatkowski DP, Swerdlow HP, et al. 2012. Optimizing illumina next-generation sequencing library preparation for extremely at-biased genomes. BMC Genomics. 13(1):1. doi:10.1186/1471-2164-13-1.
- Park JS, Kim MJ, Ahn SJ, Kim I. 2015. Complete mitochondrial genome of the grass moth Glyphodes quadrimaculalis (Lepidoptera: crambidae). Mitochondrial DNA. 26(2):247–249. doi:10.3109/19401736.2013.823183.
- Que SQ, Yu AL, Tu YG, Xiong CY, Liu XH. 2019. Complete mitochondrial genome and phylogenetic analysis of Diaphania perspectalis. Mitochondrial DNA Part B. 4(1):933–934. doi:10.1080/23802359.2019.1568213.
- Williams A, Storton D, Buckles J, Llinas M, Wang W. 2012. Improvement of PCR-free NGS Library Preparation to Obtain Uniform Read Coverage of Genome with Extremely High AT Content. J Biomolecular Techn JBT. 23(Suppl):S34.
- Yang MS, Song L, Mao JH, Shi YX, Wu CJ, Zhang YX, Huang L, Peng WF, Liu XM. 2018. Complete mitochondrial genome of the soybean leaffolder, Omiodes indicata (Lepidoptera: pyraloidea: crambidae), and phylogenetic analysis for Pyraloidea. Int J Biol Macromol. 115:53–60. doi:10.1016/j.ijbiomac.2018.03.041.
- Yang M, Shi S, Dai P, Song L, Liu X. 2018. Complete mitochondrial genome of Palpita hypohomalia (lepidoptera: pyraloidea: crambidae) and its phylogenetic implications. Eur J Entomol. 115(1):708–717.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhao J, Sun Y, Xiao LB, Tan YG, Bai LX. 2016. Complete mitochondrial genome of Cotton Leaf Roller Haritalodes derogata (Lepidoptera: crambidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2833–2834. doi:10.3109/19401736.2015.1053115.
- Zhou N, Dong Y, Qiao P, Yang Z. 2020. Complete mitogenomic structure and phylogenetic implications of the genus Ostrinia (Lepidoptera: crambidae). Insects. 11(4):232. doi:10.3390/insects11040232.