Abstract
Dendrobaena veneta (Rosa, 1886) is widely distributed all over Europe due to its use as compost worm. The specimen presented here was collected in Tiranë district, Albania. Currently, only two species’ complete or nearly complete mitochondrial genome (mitogenome) sequences have been reported in the genus Dendrobaena; D. octaedra (Savigny, 1826) and D. tellermanica Perel, 1966. In this study, the complete mitogenome of D. veneta was sequenced, assembled, and annotated. The mitogenome of D. veneta is a circular DNA molecule, consisting of 15,475 bp with an A + T content of 61.2%. It contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 non-coding region (control region). Phylogenetic analysis showed that D. veneta is clustered with the other two Dendrobaena species in the well-supported family Lumbricidae.
Introduction
The genus Dendrobaena Eisen, 1873 is one of the most speciose genera in the family Lumbricidae. Dendrobaena veneta (Rosa, 1886) is a widely distributed peregrine species, probably of East Mediterranean origin (Szederjesi et al. Citation2019; ). As it is one of the earthworms suitable for vermicomposting this species is established all over Europe, where it is mainly found in compost and manure heaps and other disturbed habitats rich in organic matter. D. veneta is a medium-sized worm with vivid red–violet stripes and the following morphological characteristics: length 30–110 mm, diameter 4–8 mm, and 54–150 segments. Clitellum on 26, 27–32, 33, tubercles on 30–31. Spermathecal pores in 9/10, 10/11 near M (Csuzdi and Zicsi Citation2003). It is closest to its sister species Dendrobaena succinta (Rosa, 1905) distributed in natural habitats of the East Mediterranean but easy to distinguish because D. succinta has a uniform red coloration and lacks the characteristic stripes of D. veneta (Szederjesi et al. Citation2019).
Materials
Samples
The specimens of D. veneta were collected in Albania, Tiranë district, Gropë Mts, Bizë, Kaprol stream, and its sidebrook at the military camp (41°33′92″N, 20°19′89″E; 1250 m) on 7 July 2019 (T. Szederjesi coll). The specimen was identified morphologically according to the characters detailed in Szederjesi et al. (Citation2019) and corroborated by using its COI barcode as well. The specimen is deposited at Jeonbuk National University (Yong Hong, [email protected]) under the voucher number JBNU0023.
Methods
Sequencing, assembling, and annotation
Total DNA was isolated from single specimen of D. veneta. A sequencing library was constructed using the Illumina TruSeq DNA Nano Library Prep Kit (Illumina Inc., San Diego, CA, USA). The mitogenome sequence was determined using the Illumina HiSeq-X platform (San Diego, CA, USA). The raw reads were de novo assembled using SPAdes version 3.13.0 (Bankevich et al. Citation2012) and NOVOplasty version 4.3.1. The depth of coverage was determined by Geneious prime implemented with Bowtine2 by mapping the total reads to the newly assembled sequence (Figure S1). The resulting complete sequences were used for the mitogenome annotation using MITOS webserver (Donath et al. Citation2019) and manually curated based on BLAST searches in the National Center for Biotechnology Information (NCBI) database.
Phylogenetic analysis
Phylogenetic analysis was conducted using 26 earthworm (23 Lumbricidae and 3 Megascolecidae) mitogenome sequences including D. veneta. These include the publicly available complete mitogenome sequences of 23 Lumbricidae species and 3 representative Megascolecidae species as outgroups. Some mitogenome sequences (CM035405, MZ857197, MZ857199, MZ857200, MZ857201) were annotated using MITOS webserver and manually curated. Although the mitogenome of D. tellermanica is not complete, it contained the entire set of 13 protein-coding genes (PCGs); therefore, it was included in the analysis as well. Nucleotide sequences of 13 PCGs from each mitogenome were extracted using Geneious Prime 2023. Each gene was aligned by the MAFFT version 7 (Katoh and Standley Citation2013). Gaps and ambiguous sites in the alignments were then trimmed using trimAl version 2.3 (Capella-Gutiérrez et al. Citation2009). Geneious Prime 2023 was used to concatenate individual gene alignments into a dataset. Substitution saturation for the 13 PCGs dataset was assessed by Xia’s test and index of substitution saturation (Iss) with a GTR model as implemented in DAMBE v7.3.32 (Xia Citation2018). The result revealed that the 13 PCGs were not saturated, with the value of the Iss obviously lower than the critical Iss value. Best-fitting nucleotide substitution model was selected on the basis of the Bayesian information criterion with ModelFinder implemented in IQ-TREE v 2.1.3 (Nguyen et al. Citation2015). An ultrafast bootstrap maximum-likelihood tree was created with IQ-TREE with 5000 bootstrap replicates using the GTR + F + I + G4 substitution model. Bayesian inference phylogeny was inferred using MrBayes v3.2.7a (Ronquist et al. Citation2012) under the GTR + I + G + F model (2 parallel runs, 1,000,000 generations), in which the initial 25% of sampled data were discarded as burn-in. Convergence was assessed by parameters including effective sample sizes (ESS) and average standard deviation of split frequencies (ASDSF). The values (ESS > 200 and ASDSF < 0.01) were considered adequate.
Results
The complete mitogenome of D. veneta is a circular DNA molecule consisting of 15,475 bp. Its structure is typical for earthworms with 13 PCGs, 22 tRNAs, and 2 rRNAs () located on the heavy strand. We observed that all 13 PCGs start with an ATG codon. Of these, 10 PCGs end with complete stop codons, TAA and TAG, and 3 PCGs end with an incomplete stop codon, T. The A + T content of the whole mitogenome is 61.2%. The sequencing reads were mapped to the assembled mitogenome sequences to evaluate the coverage of depth. At present, only a few complete Lumbricidae mitochondrial genome (mitogenome) sequences are available. Two mitogenome sequences have been reported to date, especially in the genus Dendrobaena, including a complete sequence of D. octaedra and a near complete sequence of D. tellermanica. D. veneta in the maximum-likelihood and Bayesian inference trees grouped together with the other two Dendrobaena species (), and the positions of the other Lumbricidae species are similar to the previous studies (Csuzdi et al. Citation2022; Zhao et al. Citation2022; Csuzdi et al. Citation2023; Shekhovtsov et al. Citation2023). The newly analyzed D. veneta formed a well-supported clade together with the other two Dendrobaena species including D. octaedra the type of the genus. Not surprisingly, D. veneta seems to be most close to the Caucasian D. tellermanica which corroborates the Eastern Mediterranean origin of D. veneta (Perel Citation1979; Szederjesi et al. Citation2019).
Figure 2. Circular sketch map of the Dendrobaena veneta mitogenome. The mitogenome map was generated using the GenBank format of the sequence (OQ763213) with the online Proksee software (Grant et al. Citation2023). From the inside to the outside, the innermost scale indicates the length, the inner circle indicates the GC skew, the Middle circle indicates the GC content, and the outer circle indicates gene arrangement. The plots of the GC content and GC skew were generated by default setting of the software (window 500, step 1). different colors represent different gene blocks.
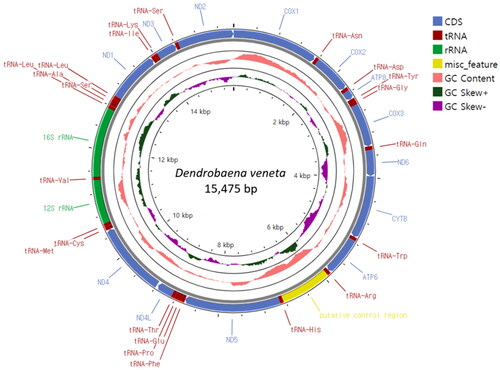
Figure 3. Phylogenetic tree of the 26 species of the Megascolecidae and Lumbricidae. Phylogenetic trees were reconstructed using maximum-likelihood (ML) and Bayesian inference (BI) methods, based on the nucleotide sequence of 13 PCGs. The sequences used in the tree are listed in . The numbers at each node specify the BI posterior probability and ultrafast bootstrap support (%), respectively. The scale bar indicates the number of substitutions per site.

Table 1. List of metagynophora mitogenomes used in this study.
Discussion and conclusions
Morphologically the genus Dendrobaena is quite heterogeneous especially because it contains species with fasciculate and pinnate types of longitudinal musculature (Csuzdi and Zicsi Citation2003) a character which was once thought to be of high importance in earthworm phylogeny (Pop Citation1941; Omodeo Citation1956; Zicsi Citation1985). However, previous mitogenome analyses have shown that this character is highly homoplasious (Shekhovtsov et al. Citation2020; Csuzdi et al. Citation2022). This observation is supported by our present studies as well, since D. veneta has fasciculate but D. octaedra possesses pinnate musculature.
However, our knowledge on Oligochaeta, especially Lumbricidae mitogenomes is still limited. More mitogenome sequences of Oligochaeta species would be helpful in creating more robust mitogenome-based phylogeny and for further understanding of Oligochaeta mitogenome evolution (Boore and Brown Citation1995).
Author contributions
Conceptualization, YH; methodology, JCK and CC; data analysis, CC, NJC, and JCK; investigation, CC, JCK, and TS; resources, TS; data curation, JCK and YH; original draft preparation, YH and JCK; review and editing, NJC and YH; project administration, YH and NJC; funding acquisition, YH. All authors have read and agreed to the published version of the manuscript.
Ethical approval
The samples used in this study are earthworms that are not included in the list of protected animals, and hence, the ethical statement is not applicable.
Supplemental Material
Download MS Word (100.6 KB)Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under accession No. OQ763213. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA769829, SRR24457999, and SAMN34844758, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Boore JL, Brown WM. 1995. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics. 141(1):305–319. doi: 10.1093/genetics/141.1.305.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348.
- Csuzdi C, Koo J, Hong Y. 2022. The complete mitochondrial DNA sequences of two sibling species of lumbricid earthworms, Eisenia fetida (Savigny, 1826) and Eisenia andrei (Bouche, 1972) (Annelida, Crassiclitellata): comparison of mitogenomes and phylogenetic positioning. Zookeys. 1097:167–181. doi: 10.3897/zookeys.1097.80216.
- Csuzdi C, Koo J, Hong Y. 2023. Complete Mitochondrial genome of Eisenia nordenskioldi pallida Malevich, 1956 from Korea, with remarks on the phylogeny of the E. nordenskiodi complex (Megadrili; Lumbricidae). Zootaxa. 5255(1):82–92. doi: 10.11646/zootaxa.5255.1.12.
- Csuzdi C, Zicsi A. 2003. Earthworms of Hungary (Annelida, Oligochaeta, Lumbricidae). In: Csuzdi C, Mahunka S, editors. Pedozoologica Hungarica. Vol. 1. Budapest (Hungary): Systematic Zoology Research Group of the Hungarian Academy of Sciences, Hungarian Natural History Museum; p. 1–271.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar V, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi: 10.1093/nar/gkad326.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Koo J, Hong Y. 2023. The complete mitochondrial genome of the Korean endemic earthworm Amynthas deogyusanensis (Clitellata: Megascolecidae). Mitochondrial DNA B Resour. 8(1):107–109. doi: 10.1080/23802359.2022.2161839.
- Liu H, Zhang Y, Xu W, Fang Y, Ruan H. 2021. Characterization of five new earthworm mitogenomes (Oligochaeta: Lumbricidae): mitochondrial phylogeny of Lumbricidae. Diversity. 13(11):580.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Omodeo P. 1956. Contributo alla revisione dei Lumbricidae. Arch Zool Ital. 41:129–212.
- Perel TS. 1979. Range and regularities in the distribution of earthworms of the USSR fauna. Moscow: Nauka. p. 272. pp. [in Russian]
- Pop V. 1941. Zur phylogenie und Systematik der Lumbriciden. Zool Jahrb Syst. 74:487–522.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Shekhovtsov SV, Peltek SE. 2019. The complete mitochondrial genome of Aporrectodea rosea (Annelida: Lumbricidae). Mitochondrial DNA B Resour. 4(1):1752–1753. doi: 10.1080/23802359.2019.1610091.
- Shekhovtsov SV, Shipova AA, Poluboyarova TV, Vasiliev GV, Golovanova EV, Geraskina AP, Bulakhova NA, Szederjesi T, Peltek SE. 2020. Species delimitation of the Eisenia nordenskioldi complex (Oligochaeta, Lumbricidae) using transcriptomic data. Front Genet. 11:598196. doi: 10.3389/fgene.2020.598196.
- Shekhovtsov SV, Vasiliev GV, Latif R, Poluboyarova TV, Peltek SE, Rapoport IB. 2023. The mitochondrial genome of Dendrobaena tellermanica Perel, 1966 (Annelida: Lumbricidae) and its phylogenetic position. Vavilovskii Zhurnal Genet Selektsii. 27(2):146–152.
- Szederjesi T, Pavlíček T, Márton O, Krízsik V, Csuzdi C. 2019. Integrative taxonomic revision of Dendrobaena veneta (Rosa, 1886) sensu lato with description of a new species and resurrection of Dendrobaena succinta (Rosa, 1905) (Megadrili: Lumbricidae). J Nat Hist. 53(5–6):301–314. doi: 10.1080/00222933.2019.1593537.
- Wang AR, Hong Y, Win TM, Kim I. 2015. Complete mitochondrial genome of the Burmese giant earthworm, Tonoscolex birmanicus (Clitellata: Megascolecidae). Mitochondrial DNA. 26(3):467–468. doi: 10.3109/19401736.2013.830300.
- Xia X. 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 35(6):1550–1552. doi: 10.1093/molbev/msy073.
- Zhang Q, Liu H, Zhang Y, Ruan H. 2019. The complete mitochondrial genome of Lumbricus rubellus (Oligochaeta, Lumbricidae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2677–2678. doi: 10.1080/23802359.2019.1644242.
- Zhao H, Fan S, Aspe NM, Feng L, Zhang Y. 2022. Characterization of 15 earthworm mitogenomes from Northeast China and its phylogenetic implication (Oligochaeta: Lumbricidae, Moniligastridae). Diversity. 14(9):714. doi: 10.3390/d14090714.
- Zicsi A. 1985. Über die Gattungen Helodrilus Hoffmeister, 1845 und Proctodrilus gen. n. (Oligochaeta: Lumbricidae). Acta Zool Hung. 31:275–289.

