Abstract
Phyla nodiflora (Linnaeus) Greene (1899) is a perennial creeping herb belonging to the family Verbenaceae. It has numerous pharmacological properties, including anti-dandruff, anti-inflammatory, anti-oxidant, anti-melanogenesis, anti-hypertensive, and anti-hyperuricemic properties. We generated the complete chloroplast genome sequence of P. nodiflora using Illumina paired-end sequencing data. The P. nodiflora chloroplast genome is 154,341 bp in length, containing a large single copy (LSC) region of 85,185 bp and a small single copy (SSC) region of 17,222 bp, separated by a pair of inverted repeats (IRs) of 25,967 bp. The genome contained 128 genes, including 86 protein-coding, 34 tRNA, and eight rRNA genes. Six genes had one intron, one gene had two introns, and the others did not have an intron. Overall GC content of the chloroplast genome was 39%, while those of LSC, SSC, and IR regions were 38.2%, 33.7%, and 44%, respectively. Phylogenetic analysis of the chloroplast genome revealed that P. nodiflora is closely related to the other species from Verbenaceae.
Introduction
The genus Phyla includes five accepted species, namely Phyla betulifolia (Kunth) Greene, Phyla cuneifolia (Torr.) Greene, Phyla lanceolata (Michx.) Greene, Phyla linearis (Kunth) Tronc. & Lopez-Pal, and Phyla nodiflora (L.) Greene (O'Leary and Múlgura Citation2012). P. nodiflora is a perennial creeping herb with obovate leaves, entire at the base and serrate at the apex. Leaves are thinly pubescent. The flowers have white or purple petals with yellow in the center. Stems are decumbent, terete, and rooting at the nodes. P. nodiflora was reported to have flavonoids (Basu et al. Citation1969), steroids (Siddiqui et al. Citation2009), volatile phytochemicals (Elakovich and Stevens Citation1985), and other compounds (Lin et al. Citation2014). The plant is mainly used as an anti-dandruff (Fatima Citation2021), anti-bacterial, anti-cancer, hepatoprotective (Arumanayagam and Arunmani Citation2015), anti-melanogenesis (Yen et al. Citation2012), anti-inflammatory (Balakrishna et al. Citation1996), and anti-hyperuricemic (Cheng et al. Citation2015) activities. In this study, we report the chloroplast genome of P. nodiflora for the first time, which provides information for further taxonomic and DNA barcoding studies in this species.
Materials and methods
Plant material of P. nodiflora was collected from wasteland and roadside at Guduvanchery, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°51′16.1″N 80°03′49.4″E; ). The voucher specimen (MH178191) was deposited in the Madras Herbarium, Botanical Survey of India (BSI), Southern Regional Centre, Coimbatore, Tamil Nadu, India, (https://bsi.gov.in/regional-centres/en?rcu=133, Dr. M. U. Sharief, Scientist-E and Head of Office, email: [email protected]). Total genomic DNA from P. nodiflora was extracted using the CTAB method (Doyle and Doyle Citation1987), with some modifications (Balaji and Parani Citation2022). According to manufacturer protocol, a whole-genome DNA sequencing library was constructed using the Nextera XT Library Prep Kit (Cat. No. FC-131-1024). The library was sequenced on the Illumina Novoseq 6000 platform (Illumina Inc., San Diego, CA, USA) with a paired-end sequencing length of 150 bp. Paired-end sequencing generated 4.68 Gb of data with q30 bases of more than 87%. The chloroplast genome assembly was done using NovoPlasty v.4.3.2 (Dierckxsens et al. Citation2017) and GetOrganelle v1.7.7.0 (Jin et al. Citation2020), with ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) gene from Nicotiana tabacum (GenBank accession no. NC_001879.2) as a seed sequence. The assembled P. nodiflora chloroplast genome was annotated with GeSeq (Tillich et al. Citation2017). The predicted transfer RNAs (tRNAs) were confirmed by tRNAscan-SE 2.0 (Lowe and Chan Citation2016). In addition, the CPGView (www.1kmpg.cn/cpgview/; Liu et al. Citation2023) was applied to structures to visualize the intron-containing genes. The sequencing depth of assembled chloroplast genome was done by aligning to the raw reads using a BWA aligner (Li and Durbin Citation2009). The bam file was viewed using the software Qualimap for coverage map (Fernando et al. Citation2012). A maximum likelihood tree of molecular distance analysis was performed using 1000 bootstrap replicates with the (GTR + G) model in MEGA version 11.0.13 (Tamura et al. Citation2021) based on the alignments created using the MAFFT program (Katoh and Standley Citation2013).
Figure 1. Phyla nodiflora plant used in this study. A. Collected sample, B. Habitat, and C. Digital herbarium (MH178191). The features that differentiate P. nodiflora from the other species of this genus are obovate leaves with sub-obtuse apex, pinnate venation prominent abaxially, serrate leaf margin, spherical-shaped fruit and rooting at the nodes (O'Leary and Múlgura Citation2012). These images were taken by authors (Ray Sharmishtha and Tanuja). the plant sample was collected from Guduvanchery, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°51′16.1″N 80°03′49.4″E) on 9 September 2022.

Results and discussion
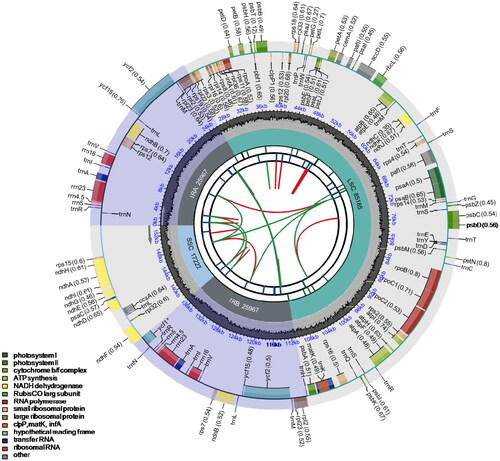
The chloroplast genome of P. nodiflora was 154,341 bp with a mean organelle coverage of 378× (). The chloroplast genomes assembled using NOVOPlasty and GetOrganelle were 100% identical. It had a typical quadripartite structure, including a large single copy (LSC) region of 85,185 bp, a small single copy (SSC) region of 17,222 bp, and a pair of inverted repeats (IRs) of 25,967 bp. The chloroplast genome contained 86 protein-coding genes, 34 tRNA genes, and eight rRNA genes. Six genes (atpF, ndhA, ndhB, rpl2, rpoC1, rps16) were single-intron genes, and one gene (pfaI) had two introns (). A Chloroplast genome of comparable size (154,310 bp) was reported from Lippia origanoides, which is closely related to P. nodiflora. L. origanoides contained the same number of rRNA genes, but the number of protein-coding genes (88) and tRNA genes differed in small numbers compared to P. nodiflora (Sarzi et al. Citation2019). This comparative analysis highlights the structural similarities and differences between these closely related species within the broader context of phylogenetic relationships. The overall GC content was 39%, while LSC, SSC, and IR regions were 38.2%, 33.7%, and 44%, respectively. The average sequencing depth of the bases in the assembled genome was between 52× and 5644×, with a mean of 2041.3× (Supplementary Figure 3). The complete chloroplast genome of P. nodiflora with annotations was submitted to GenBank (Accession No. OQ673174). The raw reads were deposited in the GenBank Sequence Read Archive (Accession No. SRR23875832).
Figure 2. Circular map of the chloroplast genome of Phyla nodiflora. From the center going outward, the first circle shows the distribution of the repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle displays the tandem repeats marked with short bars. The third circle shows the LSC, SSC, IRa, and IRb regions. The fourth circle shows the percentage of GC content. The next circle shows the genes having different colors based on the functional groups. The functional classification is shown at the bottom left. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counter-clockwise direction.
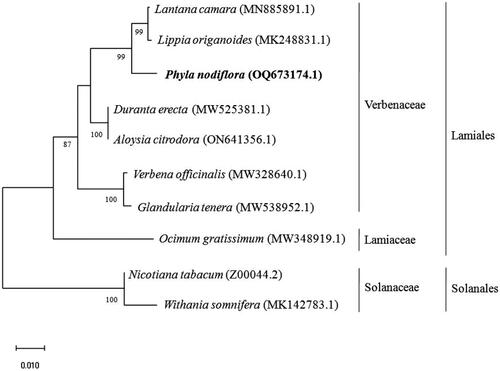
To confirm the phylogenetic positions of P. nodiflora in the Verbenaceae, the complete chloroplast genome of P. nodiflora (OQ673174) and the other species of the Verbenaceae family, namely Duranta erecta (MW525381.1), Aloysia citrodora (ON641356.1), Lantana camera (MN885891.1), Lippia origanoides (MK248831.1), Glandularia tenera (MW538952.1), and Verbena officinalis (MW328640.1) were downloaded from NCBI database. Ocimum gratissimum (MW348919.1) from Lamiaceae, Nicotiana tabacum (Z00044.2), and Withania somnifera (MK142783.1) from the Solanaceae family were used as outgroups. The phylogenetic analysis fully resolved P. nodiflora in a clade with Lantana camera and Lippia origanoides within the Verbenaceae family (). A phylogenetic tree constructed based on the polymorphisms in ETS, ITS, trnL/rpl32, PHOTII, and seven pentatrichopeptide repeat (PPR) genes for the members of Lantana, Lippia, Aloysia, Nashia, Phyla, Burroughsia, and Acantholippia genera of Verbanacea also showed that Phyla, Lippia, and Lantana were closely related (Lu‐Irving et al. Citation2021). The complete chloroplast genome of P. nodiflora can be subsequently used for phylogenetic analysis, DNA barcoding, and molecular marker studies to authenticate the herbal product of P. nodiflora.
Figure 3. Phylogenetic tree constructed by maximum likelihood analysis based on complete chloroplast genome sequences, including the chloroplast genome of P. nodiflora (OQ673174). The bootstrap support values are shown on the nodes. The chloroplast genome sequences used for the phylogenetic analysis were Ocimum gratissimum MW348919.1 (Balaji et al. Citation2021), Duranta erecta MW525381.1 (Song et al. Citation2021), Aloysia citrodora ON641356.1, Verbena officinalis MW328640.1 (Yue et al. Citation2021), Phyla nodiflora OQ673174.1, Lanata camera MN885891.1, Lippia origanoides MK248831.1 (Sarzi et al. Citation2019), Nicotiana tabacum Z00044.2 (Shinozaki et al. Citation1986), and Withania somnifera MK142783.1 (Mehmood et al. Citation2020).
Conclusion
This study assembled and annotated the complete chloroplast sequence of Phyla nodiflora for the first time. P. nodiflora and other species were entirely resolved in a clade by the phylogenetic analysis within the Verbenaceae family. Following that, phylogenetic analysis, DNA barcoding, and molecular marker studies can be used to confirm the authenticity of the herbal products derived from P. nodiflora.
Ethical approval
No permissions were required for the sample collection of Phyla nodiflora L. because it is widely distributed in the wastelands and roadsides in tropical regions. The plant species was collected from Guduvanchery, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°51′16.1″N 80°03′49.4″E).
Author contributions
Ray Sharmishtha, Tanuja, and R. Balaji collected the specimen material, conducted the experiment, analyzed the sequence data, and drafted the paper. M. Parani contributed to the conception and design of this work. All the authors carefully read, revised, and approved the final manuscript to be published. We thank Dr. Senthilkumar Umapathy for his help in the authentication of plant material.
Supplemental Material
Download TIFF Image (74.2 MB)Supplemental Material
Download TIFF Image (2.9 MB)Supplemental Material
Download TIFF Image (9.2 MB)Disclosure statement
The authors reported no potential competing interests.
Data availability statement
The genome sequence data supporting the findings of this study are available in NCBI under the accession number OQ673174 (https://www.ncbi.nlm.nih.gov/nuccore/OQ673174.1). The associated next-generation sequencing data files are available from the BioProject, Bio-Sample, and SRA submission under the accession numbers PRJNA945246, SAMN33776799, and SRR23875832, respectively.
Additional information
Funding
References
- Arumanayagam S, Arunmani M. 2015. Hepatoprotective and antibacterial activity of Lippia nodiflora Linn. against lipopolysaccharides on HepG2 cells. Pharmacogn Mag. 11(41):24–31. doi: 10.4103/0973-1296.149689.
- Balaji R, Parani M. 2022. DNA barcoding of the market samples of single‐drug herbal powders reveals adulteration with taxonomically unrelated plant species. Diversity. 14(6):495. doi: 10.3390/d14060495.
- Balaji Raju, Ravichandiran Kumar, Parani Madasamy, Tanuja . 2021. The complete chloroplast genome of Ocimum gratissimum from India – a medicinal plant in the Lamiaceae.Mitochondrial DNA B Resour. 6(3):948–950. doi: 10.1080/23802359.2021.1889413.
- Balakrishna K, Gopal RH, Ramkumar V, Rao RB, Vasanth S, Narayanappa D. 1996. Antibacterial activity of the essential oil of Lippia nodiflora. Anc Sci Life. 16(1):79.
- Basu AK, Chakraborti P, Sanyal PK. 1969. Nodifloretin – a new flavone from Lippia nodiflora. J Indian Chem Soc. 46(4):271–272.
- Cheng LC, Murugaiyah V, Chan KL. 2015. Flavonoids and phenylethanoid glycosides from Lippia nodiflora as promising anti-hyperuricemic agents and elucidation of their mechanism of action. J Ethnopharmacol. 176:485–493. doi: 10.1016/j.jep.2015.11.025.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Elakovich SD, Stevens KL. 1985. Volatile constituents of Lippia nodiflora. J Nat Prod. 48(3):504–506. doi: 10.1021/np50039a037.
- Fatima SA. 2021. Formulation of poly herbal hair oils and evaluation of its anti-dandruff activity against the fungus Malassezia (Malassezia furfur). Int J Res Appl Sci Eng Technol. 9(VI):4106–4110. doi: 10.22214/ijraset.2021.35899.
- Fernando G, Konstantin O, José C, Luis MC, Stefan G, Sonia T, Joaquín D, Thomas FM, Ana C. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 28(20):2678–2679.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324.
- Lin FJ, Yen FL, Chen PC, Wang MC, Lin CN, Lee CW, Ko HH. 2014. HPLC-fingerprints and antioxidant constituents of Phyla nodiflora. Sci World J. 2014:528653–528658. doi: 10.1155/2014/528653.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi: 10.1093/nar/gkw413.
- Lu‐Irving P, Bedoya AM, Salimena FR, dos Santos Silva TR, Viccini LF, Bitencourt C, Thode VA, Cardoso PH, O'Leary N, Olmstead RG. 2021. Phylogeny of Lantana, Lippia, and related genera (Lantaneae: Verbenaceae). Am J Bot. 108(8):1354–1373. doi: 10.1002/ajb2.1708.
- Mehmood F, Abdullah Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2):1522–1530. doi: 10.1016/j.ygeno.2019.08.024.
- O'Leary N, Múlgura ME. 2012. A taxonomic revision of the genus Phyla (Verbenaceae). Ann Mo Bot Gard. 98(4):578–596. doi: 10.3417/2009120.
- Sarzi DS, Haerolde L, Lopes FS, Furtado C, Oliveira DR, Sakuragui CM, Prosdocimi F. 2019. Complete plastid genome of Lippia origanoides (Verbenaceae) and phylogenomic analysis of Lamiales. Mitochondrial DNA B: Resour. 4(1):808–810. doi: 10.1080/23802359.2019.1574675.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x.
- Siddiqui BS, Ahmed F, Ali SK, Perwaiz S, Begum S. 2009. Steroidal constituents from the aerial parts of Lippia nodiflora Linn. Nat Prod Res. 23(5):436–441. doi: 10.1080/14786410802044802.
- Song WC, Feng Q, Zhang YJ, Wu XM, Shi C, Wang S. 2021. The complete chloroplast genome sequence of Duranta erecta (Verbenaceae). Mitochondrial DNA B Resour. 6(7):1832–1833. doi: 10.1080/23802359.2021.1934164.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Yen FL, Wang MC, Liang CJ, Ko HH, Lee CW. 2012. Melanogenesis inhibitor(s) from Phyla nodiflora extract. Evid Based Complement Alternat Med. 2012;867494. doi: 10.1155/2012/867494.
- Yue Z, Wang J, Wang Y, Zhou B, Zhou X, Zhang W, Li C, Qi Z, Wang H. 2021. The complete chloroplast genome of Verbena officinalis, an herbal species of Verbenaceae family. Mitochondrial DNA B Resour. 6(7):1982–1983. doi: 10.1080/23802359.2021.1938719.
