Abstract
The genus Allium comprises some of the most commonly consumed food crops worldwide. The chloroplast genomes of A. sacculiferum, A. thunbergii, and A. taquetii are 152,444, 153,459, and 154,056 bp circular molecular genomes, respectively. The annotation results revealed the presence of 132 (89 protein-coding, 35 tRNA, and eight rRNA), 132 (86 protein-coding, 38 tRNA, and eight rRNA), and 132 (86 protein-coding, 38 tRNA, and eight rRNA) genes with 36.78%, 36.83%, and 36.88% total GC content in each genome, respectively. The chloroplast genome of each Allium species contains an 81,254, 82,473, and 83,068 bp LSC region, an 18,176, 18,006, and 17,958 bp SSC region, and a pair of 26,507, 26,490, and 26,515 bp IR regions, respectively. Phylogenetic analysis based on the 21 complete chloroplast genomes indicates that A. sacculiferum is closely related to A. koreanum; A. thunbergii and A taquetii are closely related to A. hookeri. This study shows that the three Allium species are Korean crop wild relatives that may be used to develop new Allium varieties in the future.
Introduction
Most food resources can be obtained from wild plants; hence, research on crop wild relatives (CWRs) is essential for the conservation of genetic diversity in crops. Therefore, collection, research, and conservation of CWRs are being performed worldwide as essential research in the context of food security to conserve and increase crop species diversity (Global Crop Wild Relatives Project; GCWR).
Allium L. is a genus within the family Amaryllidaceae that comprises more than 800 species including garlic and onion (Li et al. Citation2010; Deniz et al. Citation2015). Of these, A. sacculiferum Max., A. thunbergii G. Don, and A. taquetii H. Lév. & Vaniot are native to Korea (Choi and Oh Citation2011). A. sacculiferum Max. (Maxim, 1887) is a perennial herb with thicker and longer leaves than those of the common leek (Lee and Lee Citation1997). The root produces 2–3 leaves, the lower part of which surrounds the flower stem. The plant produces purple red flowers that bloom between July and September (Lee Citation2006). A. thunbergii G. Don (Don, 1827) is a perennial herb that is 30–60 cm in height and prevalent in mountainous areas. It flowers between August and September and has 2–3 leaves that are 2–5 mm in diameter, spread obliquely upward, and whitish green in color with triangular cross-section (Lee Citation2006). A. taquetii H. Lév. & Vaniot (Fedde, 1908) is a wild perennial herb that grows in rock crevices at an altitude of 1100 m in Hallasan, Jirisan, and Gayasan mountains. It has 3–4 leaves that are 15–20 cm long, 2–3 mm wide, and smaller than the flower stem, similar to leek leaves. It has purple flowers that bloom between September and October (Lee Citation2006; Guh et al. Citation2015).
The present study aimed to assemble the complete chloroplast genome of A. sacculiferum Maxim., A. thunbergii G. Don, and A. taquetii H. Lév. & Vaniot to provide genetic sources for further research. Additionally, we attempted to establish CWRs of Korean Allium species of high commercial value using their chloroplast genomes.
Materials and methods
The three Allium species were provided by the Baekdudaegan National Arboretum seed bank (A. sacculiferum: B0007205, A. thunbergii: B0015888, and A. taquetii: B0004146/Contact: Kyeong Min Kim, [email protected]) (). A. sacculiferum seeds were collected from Sambongsan Mountain, Bongsan-ri, Goje-myeon, Geochang-gun, Gyeongsangnam-do (35°53′01.0″N 127°51′09.7″E). A. thunbergii seeds were collected from Hyangnobong Peak, Jinbu-ri, Ganseong-eup, Goseong-gun, Gangwon-do (38°19′59.9″N 128°19′06.2″E). A. taquetii seeds were collected from the Baekdudaegan National Arboretum (37°00′23.0″N 128°48′36.7″E). Voucher specimens were deposited in the Baekdudaegan National Arboretum (A. sacculiferum: 2019-008952, A. thunbergii: 2020-040355, A. taquetii: 2017-006634/Contact: Kyeong Min Kim). Post-germination fresh leaves were used to extract DNA using a modified cetyl trimethylammonium bromide protocol (Doyle and Doyle Citation1987).
Figure 1. Images of the Allium species. Photographs of the reference images were taken by the authors at the Baekdudaegan National Arboretum, South Korea, without any copyright issues. Seeds were provided by Baekdudaegan National Arboretum Seed bank and propagated in the Baekdudaegan National Arboretum. (A) A. sacculiferum Maxim. (B) A. thunbergii G. Don. (C) A. taquetii H. Lév. & Vaniot.

The sequencing library was produced using the QIAseq FX DNA library CDI kit (Qiagen, Hilden, Germany), which was constructed with an insert length of 350 bp and paired-end sequenced with 151 bp reads. The prepared library was loaded onto the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA), and the reads were subsequently filtered and trimmed using the FASTX-Toolkit. The trimmed reads were then used to assemble the complete chloroplast genome using the GetOrganelle Pipeline (Jin et al. Citation2020). The genomes of A. sacculiferum Max, A. thunbergii G. Don, and A. taquetii were annotated using AGORA (Jung et al. Citation2018) by comparing the chloroplast genome sequences of Allium chinense (NCBI accession number: NC_043922.1), Allium senescens (NCBI accession number: NC_057580.1), and Allium tuberosum (NCBI accession number: MN158715.1). The circular chloroplast genome map was generated using OGDRAW ver 1.3.1 (Greiner et al. Citation2019). IRscope was used to compare the boundaries of four chloroplast genome regions, namely large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRa and IRb) (Amiryousefi et al. Citation2018). The schematic map of the cis- and trans-splicing gene of the chloroplast genome was generated using CPGView (Liu et al. Citation2023, http://www.1kmpg.cn/cpgview/)
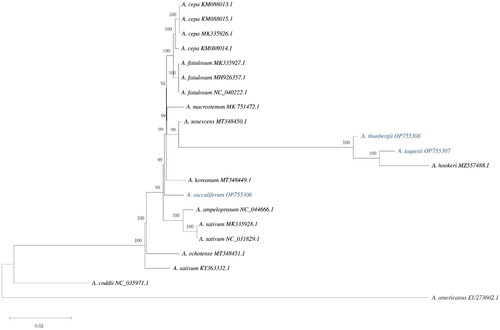
The complete chloroplast genome sequences of the three species sequenced in the current study were aligned with that of 11 species downloaded from the NCBI GenBank database using ClustalW2 (Larkin et al. Citation2007). For phylogenetic analysis, we used Agapanthus coddii, which is closely related to the genus Allium, and Acorus americanus, which was used as an outgroup in previous Allium genus studies (Huo et al. Citation2019). The maximum-likelihood (ML) method was used to construct a phylogenetic tree based on 21 complete chloroplast genomes. The ML phylogenetic analysis was performed using the MEGA 11 with 1000 bootstrap values replicate and General Time Reversible model (Tamura et al. Citation2021) ().
Figure 2. The phylogenetic tree reconstructed using MEGA based on 21 complete chloroplast genome sequences inferred using the maximum-likelihood (ML) method. All sequences were downloaded from NCBI GenBank. The sequences used for phylogenetic tree construction are as follows: A. cepa (MK335926.1; Huo et al. Citation2019/KM088013.1, KM088014.1, KM088015.1; Kim et al. Citation2015), A. sativum (NC_031829.1/KY363332.1/MK335928.1; Huo et al. Citation2019), A. fistulous (MK335927.1; Huo et al. Citation2019/NC_040222.1; Kim et al. Citation2015/MH926357.1; Yusupov et al. Citation2019), A. koreanum (MT348449.1; Namgung et al. Citation2021), A. ochotense (MT348451.1; Namgung et al. Citation2021), A. senescens (MT348450.1; Namgung et al. Citation2021), A. hookeri (MZ557488.1; Ren et al. Citation2022), A. macrostemon (MK75472.1; Xie et al. Citation2019), A. ampeloprasum (NC_044666.1), Agapanthus coddii (NC_035971.1), and Acorus americanus (EU273602.1). The numbers above the nodes indicate bootstrap support values (with 1000 replicates). The numbers under the nodes indicate the length of each node.
Results
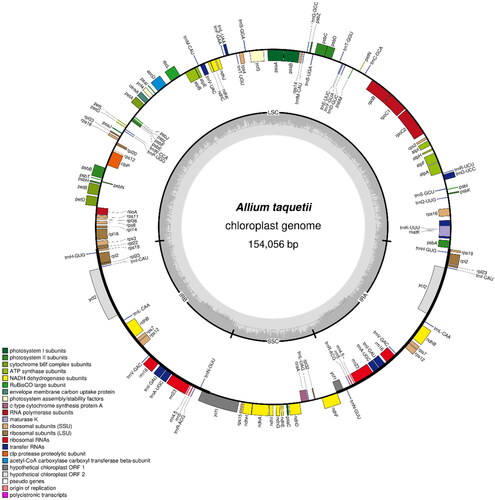
The chloroplast genomes of A. sacculiferum (NCBI accession number: OP755306), A. thunbergii (NCBI accession number: OP755308), and A. taquetii (NCBI accession number: OP755307) were found to be circular molecules that are 152,444, 153,459, and 154,056 bp long, respectively. The coverage depth of each species is shown in Figure S1. The annotation results revealed the presence of 132 (89 protein-coding, 35 tRNA, and eight rRNA), 132 (86 protein-coding, 38 tRNA, and eight rRNA), and 132 (86 protein-coding, 38 tRNA, and eight rRNA) genes with total GC contents of 36.78%, 36.83%, and 36.88% in each genome, respectively (). The chloroplast genome of each Allium species contains an LSC region that is 81,254, 82,473, and 83,068 bp, an SSC region that is 18,176, 18,006, and 17,958 bp, and a pair of IR regions that are 26,507, 26,490, and 26,515 bp long, respectively (Figure S2). In the case of A. sacculiferum, 13 genes (atpF, rpoC1, petB, rpl16, rpl2, ndhB, ndhA, trnI-GAU, and trnA-UGC) including copy genes contained one intron. In the case of the other two species, 18 genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, ndhA, trnK-UUU, trnG-UCC, trnV-UAC, trnI-GAU, trnA-UGC) including copy genes contained one intron. In all three species, rps12, ycf3, and clpP contained two introns. The structures of the cis- and trans-splicing genes are shown in Figures S3–S5.
Figure 3. Chloroplast genome map of A. sacculiferum. The center of the map indicates the species name and length of the chloroplast genome. The outer circle represents the genes, and the inner circle represents the size of the IRs, LSC, and SSC, respectively. Genes with diverse functions are shown in different colors. The dark grey plot of the inner circle represents the GC content, and the grey line in middle represents the 50% threshold.
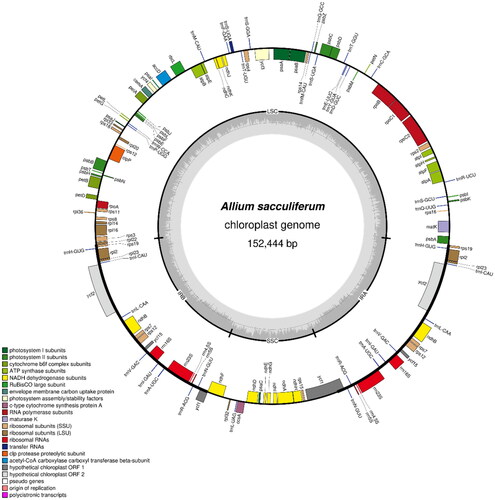
Figure 4. The chloroplast genome map of A. thunbergii. The center of the map indicates the species name and length of the chloroplast genome. The outer circle represents the gene, and the inner circle represents the size of the IRs, LSC, and SSC, respectively. Genes with diverse functions are shown in different colors. The dark grey plot of the inner circle represents the GC content, and the grey line in middle represents the 50% threshold.
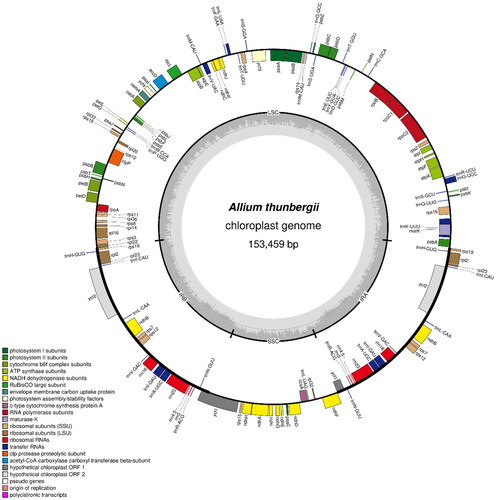
Figure 5. The chloroplast genome map of A. taquetii. The center of the map indicates the species name and length of the chloroplast genome. The outer circle represented the genes, and the inner circle represented the size of the IRs, LSC, and SSC, respectively. Genes with diverse functions are shown in different colors. The dark grey plot of the inner circle represents the GC content, and the grey line in middle represents the 50% threshold.
Most nodes in the phylogenetic tree had 99–100% bootstrap values. The phylogenetic tree revealed that all Allium species formed one monophyletic clade, and A. sacculiferum is a part of the sister group of A. koreanum. Additionally, A. thunbergii and A. taquetii were most closely related to A. hookeri with high bootstrap support. A. taquetii was supported as part of the sister group of A. hookeri.
Discussion and conclusions
The genome size (152,444–154,056 bp), overall GC content (>36%), and gene composition (protein-coding, tRNA, and rRNA) of the three Allium species showed high similarity to those of other Allium chloroplast genomes (Huo et al. Citation2019; Xie et al. Citation2020). These results suggest that the chloroplast genome of Allium has been evolutionarily conserved at the genus level. Phylogenetic analysis of 14 species showed that A. thunbergii and A. taquetii are sisters to A. hookeri. A. sacculiferum and A. thunbergii are internationally known CWRs of Allium (GCWR), and this has been verified through the phylogenetic analysis performed in this study. Additionally, the high support values warrant the inclusion of A. taquetii – a Korean endemic plant – in the list of Allium CWRs. Significantly, the complete chloroplast genomes of three Korean Allium species have been reported in this study for the first time, providing valuable data for further phylogenetic studies of Amaryllidaceae and understanding the Allium genome. Furthermore, the CWRs identified for Allium may be used beneficially in future crop breeding programs.
Author contributions
Kyeong Min Kim: methodology, data analysis, software, and writing; Chung Youl Park: conceptualization, methodology, and software; Da Hyun Lee: sample preparing and acquisition of data; Young Ho Jung: sample preparing and acquisition of data; Hyeon Min Kim: sample preparation and acquisition of data; Chae Sun Na: supervision of the project, conceptualization, methodology, writing, revision, and editing.
Ethics statement
Seed collection was performed in accordance with the guidelines and regulations provided by the Baekdudaegan National Arboretum. There are no ethical issues or other conflicts of interest in this study.
Supplemental Material
Download MS Word (1.3 MB)Disclosure statement
There are no potential conflicts of interest to disclose.
Data availability statement
The genome sequence data of Allium sacculiferum, A. thunbergii, and A. taquetii that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under accession nos. OP755306, OP755308, and OP755307, respectively. The BioProject, SRA, and Bio-Sample numbers for A. sacculiferum are PRJNA822879, SRR18707531, and SAMN27281643, respectively; for A. thunbergii are PRJNA822879, SRR18707530, and SAMN27281644, respectively; and for A. taquetii are PRJNA822879, SRR18707529, and SAMN27281645, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031. doi: 10.1093/bioinformatics/bty220.
- Choi HJ, Oh BU. 2011. A partial revision of Allium (Amaryllidaceae) in Korea and north-eastern China. Bot J Linn Soc. 167(2):153–211. doi: 10.1111/j.1095-8339.2011.01166.x.
- Deniz İG, Genç İ, Sarı D. 2015. Morphological and molecular data reveal a new species of Allium (Amaryllidaceae) from SW Anatolia, Turkey. Phytotaxa. 212(4):283–292. doi: 10.11646/phytotaxa.212.4.4.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Guh J, Kim C, Oh C, Kuk Y, Kwon O, Park K, Lee S. 2015. The natural benefits of Korean Flora as for food relief and health care. Seoul, South Korea: Resource Plant Research Group; p. 428–431.
- Huo Y, Gao L, Liu B, Yang Y, Kong S, Sun Y, Yang Y, Wu X. 2019. Complete chloroplast genome sequences of four Allium species: comparative and phylogenetic analyses. Sci Rep. 9(1):12250. doi: 10.1038/s41598-019-48708-x.
- Jin J-J, Yu W-B, Yang J-B, Song Y, DePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Jung J, Kim JI, Jeong Y-S, Yi G. 2018. AGORA: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics. 34(15):2661–2663. doi: 10.1093/bioinformatics/bty196.
- Kim S, Park JY, Yang T. 2015. Comparative analysis of the complete chloroplast genome sequences of a normal male-fertile cytoplasm and two different cytoplasms conferring cytoplasmic male sterility in onion (Allium cepa L.). J Hortic Sci Biotechnol. 90(4):459–468. doi: 10.1080/14620316.2015.11513210.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948. doi: 10.1093/bioinformatics/btm404.
- Lee Y, Lee W. 1997. Illustrated rare and endangered species in Korea. Pocheon, South Korea: Korea National Arboretum; p. 41.
- Lee TB. 2006. Coloured flora of Korea. Vol. 2. Seoul,South Korea: Hyangmunsa; p. 703–706.
- Li Q-Q, Zhou S-D, He X-J, Yu Y, Zhang Y-C, Wei X-Q. 2010. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann Bot. 106(5):709–733. doi: 10.1093/aob/mcq177.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Namgung J, Do HDK, Kim C, Choi HJ, Kim JH. 2021. Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae). Sci Rep. 11(1):3262. doi: 10.1038/s41598-021-82692-5.
- Ren F, Wang L, Zhuo W, Chen D, Huang H, Zhang L. 2022. Complete chloroplast genome of the rare medicinal vegetable Allium hookeri. Mitochondrial DNA B Resour. 7(1):6–7. doi: 10.1080/23802359.2021.2003262.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Xie FM, Jiang QP, Yu Y, Zhou SD, He XJ. 2019. The complete chloroplast genome of Allium macrostemon. Mitochondrial DNA Part B. 4(1):1938–1939. doi: 10.1080/23802359.2019.1616626.
- Xie FM, Xie DF, Xie C, Yu Y, Zhou SD, He XJ. 2020. Adaptation evolution and phylogenetic analyses of species in Chinese Allium section Pallasia and related species based on complete chloroplast genome sequences. Biomed Res Int. 2020:8542797. doi: 10.1155/2020/8542797.
- Yusupov Z, Deng T, Liu C, Lin N, Tojibaev K, Sun H. 2019. The complete chloroplast genome of Allium fistulosum. Mitochondrial DNA Part B. 4(1):489–490. doi: 10.1080/23802359.2018.1545532.
