Abstract
Swertia divaricata Harry Sm., 1965, (Gentianaceae) is a perennial herb endemic to Northwest Yunnan, China, belonging to the species-rich genus Swertia. It possesses unique morphological features but its systematic position remains uncertain. To determine its phylogenetic placement, the complete plastid genome of S. divaricata was assembled utilizing high-throughput sequencing data. The genome is circular, spanning 152,073 bp, and comprises a large single-copy (LSC) region of 82,470 bp, a small single-copy (SSC) region of 18,153 bp, and two inverted repeats (IR) regions, each 25,725 bp. A total of 130 genes were annotated, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The plastome of S. divaricata exhibits a structure and gene composition highly similar to those of other Swertia plastomes. Phylogenetic analysis indicated that S. divaricata is closely related to S. erythrosticta, sister to a subclade comprising species from sections Swertia and Apterae. The plastome sequence described herein constitutes a valuable contribution to phylogenetic and evolutionary research on Swertia.
Introduction
The genus Swertia L. (Gentianaceae) is globally distributed and comprises approximately 170 species, with the Qinghai-Tibet Plateau serving as the center of its distribution and diversity (Ho and Liu Citation2015). Among the 11 sections in Swertia, section Swertia represents one of the most species-rich taxa and encompasses 19 species. Swertia is considered one of the most taxonomically challenging genera within Gentianaceae due to paraphyletic characteristics, and the phylogenetic relationships within the genus remain contentious (Xi et al. Citation2014; Sun and Fu, Citation2019; Zhang et al. Citation2020a; Cao et al. Citation2022; Chen et al. Citation2023).
Swertia divaricata Harry Sm., 1965, a member of section Swertia, is highly valued in Chinese traditional medicine (Brahmachari et al. Citation2004; Ho and Liu Citation2015). Swertia. divaricata is endemic to Northwest Yunnan, China, and is only observed in its type locality at present (Ho and Liu Citation2015). The species exhibits distinct morphological traits within the genus, for example strictly divaricate inflorescence, 5-merous flowers, 1 to a few fimbriae at the base of the nectary. As it has not been previously incorporated into any molecular phylogenetic analyses, its systematic placement remains ambiguous. In this study, we characterize the full plastome of S. divaricata and reevaluate its phylogenetic position within the genus.
Materials and methods
We collected fresh leaves and specimens of S. divaricata () from Gongshan Town, Yunnan Province, China (28.24°N, 98.27°E). S. divaricata is not an endangered species, and specimen collection did not require permission. Dr. Pengcheng Fu identified and deposited the voucher specimen (Fu2018063) at the Herbarium of Luoyang Normal University (Bin Cai, [email protected]). We extracted total DNA using the Dzup plant genomic DNA extraction kit (Sangon, Shanghai, China). We constructed a paired-end library with an insert size of 150 bp and sequenced it using the Illumina HiSeq 2500 platform (Novogene, Tianjin, China). We filtered and trimmed raw reads with Trimmomatic v0.32 (Bolger et al. Citation2014) to eliminate adaptor sequences, low-quality reads, and sites, and checked for quality using FastQC v0.11.2. We assembled the plastome using GetOrganelle version 1.7.1 (Jin et al. Citation2020) and annotated it using GeSeq (Tillich et al. Citation2017) with default parameters. The reads coverage was plotted using a Python script (Ni et al. Citation2023). We drew the genome map using CPGview (http://www.1kmpg.cn/cpgview). We compared the plastome with Swertia species (Zhang et al. Citation2020a; Cao et al. Citation2022) and detected structural changes using mVISTA (Frazer et al. Citation2004). To confirm the phylogenetic position of S. divaricata, we extracted shared protein-coding genes from all available Swertia plastomes in PhyloSuite version 1.1.15 (Zhang et al. Citation2020b). We aligned the genes using MAFFT version 7 (Katoh and Standley Citation2013) and concatenated them to conduct maximum likelihood phylogenetic analyses with IQ-TREE version 1.6.12 (Nguyen et al. Citation2015) using 1000 replicates. We selected the best substitution model using ModelFinder 2 (Kalyaanamoorthy et al. Citation2017), and Gentiana handeliana (GenBank accession no. MN199143, Fu et al. Citation2021) was used as the outgroup.
Results and discussions
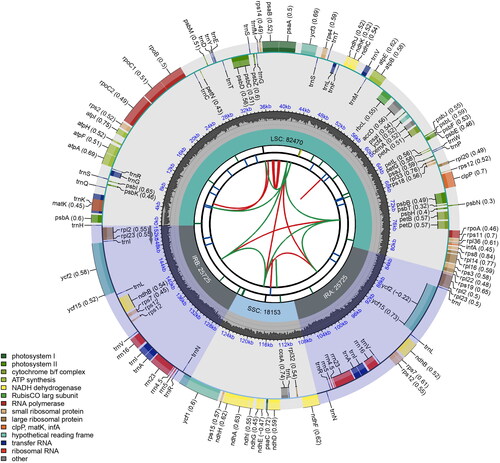
The complete plastome of S. divaricata (OQ446461) is 152,073 bp long. The LSC and SSC regions are 82,471 and 18,153 bp long, respectively, and two IRs of 25,725 bp each are present. The sequencing depth ranged from 414× to 1928× and averaged at 1368× (Figure S1). We annotated a total of 130 genes, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes (). The 18 genes in the IR are duplicated, including 7 protein-coding genes, seven tRNA genes, and four rRNA genes (). A total of 12 cis-splicing genes and one trans-splicing gene were identified (Figure S2). Comparative analysis showed that S. divaricata’s plastome structure is very similar to that in Swertia, and has no genes missing such as the ndh complex found in the subtribe Gentianinae (Sun et al. Citation2018; Fu et al. Citation2021, Citation2022).
Figure 2. Schematic map of the chloroplast genome of Swertia divaricata generated using CPGview. From center outwards, first circle shows distributed repeats connected with red (forward direction) and green (reverse direction) arcs. Second circle shows tandem repeats marked with short bars. Third circle shows microsatellite sequences as short bars. Forth circle shows sizes of LSC: long single copy, SSC: short single copy regions and inverted repeat (IRa and IRb) region. Fifth circle shows GC contents along the genome. Sixth circle shows genes marked with different colors according to their functional groups.
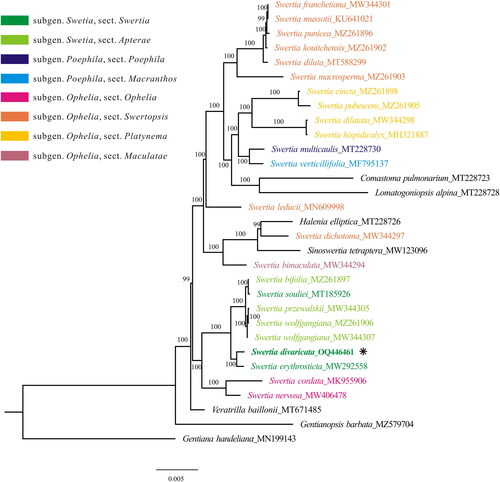
Our phylogenetic analysis using a matrix of 75 protein-coding genes revealed that Swertia is undoubtedly paraphyletic, mixing with species from the genera Comastoma, Lomatogoniopsis, Halenia, and Sinoswertia (). The backbone of our phylogenetic tree is consistent with that of Cao et al. (Citation2022), and again shows the uncertainty of the generic boundary of the genus Swertia and its allies. One reason for the phylogenetic complexity may be the ancient origin of Swertia and its allies revealed by 1280 single-copy nuclear genes (Chen et al. Citation2023). Furthermore, our analysis indicated that S. divaricata is closely related to S. erythrosticta and is the sister clade to the clade containing species from sections Swertia and Apterae, such as S. bifolia, S. przewalskii, S. wolfgangiana, and S. souliei (). Although S. souliei (section Swertia) is robustly clustered with section Apterae by our plastome data as well as single-copy nuclear genes (Chen et al. Citation2023), section Apterae including S. souliei is still not a monophyletic group due to chaos from species such as S. petiolate (Chen et al. Citation2023). Therefore, genomic data from more species shall be generated to explore the phylogenetic relationship in Swertia. This newly reported plastome sequence of S. divaricata provides valuable molecular data for illuminating the phylogenetic and molecular evolution of Swertia.
Figure 3. The maximum likelihood tree of Swertia, constructed using protein-coding genes from the plastome. Numbers above/below branches present bootstrap support. Species shown in bold is the newly sequenced in this study. The newly sequenced species was marked with an asterisk (*). The sectional classification of Swertia were marked with variable colors according to the treatment in Ho and Liu (Citation2015). the following sequences were used: S. franchetiana MW344301 (Xu et al. Citation2021), S. mussotii KU641021 (Xiang et al. Citation2016), S. punicea MZ261896 (Cao et al. Citation2022), S. kouitchensis MZ261902 (Cao et al. Citation2022), S. diluta MT588299 (Yang et al. Citation2020a), S. macrosperma MZ261903 (Cao et al. Citation2022), S. cincta MZ261898 (Cao et al. Citation2022), S. pubescens MZ261905 (Cao et al. Citation2022), S. dilatata MW344298 (Xu et al. Citation2021), S. hispidicalyx MH321887 (unpublished), S. multicaulis MT228730 (Zhang et al. Citation2020a, Citation2020b), S. verticillifolia MF795137 (Zhang et al. Citation2020a, Citation2020b), S. leducii MN609998 (unpublished), S. dichotoma MW344297 (Xu et al. Citation2021), S. bimaculata MW344294 (Xu et al. Citation2021), S. bifolia MZ261897 (Cao et al. Citation2022), S. souliei MT185926 (Bi et al. Citation2020), S. przewalskii MW344305 (Xu et al. Citation2021), S. wolfgangiana MZ261906 (Cao et al. Citation2022) MW344307 (Xu et al. Citation2021), S. erythrosticta MW292558 (unpublished), S. cordata MK955906 (Huang et al. Citation2019), S. nervosa MW406478 (Xu et al. Citation2021), Comastoma pulmonarium MT228723 (Zhang et al. Citation2020a, Citation2020b), Lomatogoniopsis alpina MT228728 (Zhang et al. Citation2020a, Citation2020b), Halenia elliptica MT228726 (Zhang et al. Citation2020a, Citation2020b), Sinoswertia tetraptera MW123096 (Yang et al. Citation2020b), veratrilla baillonii MT671485 (unpublished), Gentianopsis barbata MZ579704 (Feng et al. Citation2022), Gentiana handeliana MN199143 (Fu et al. Citation2022).
Ethical approval
The materials used in this study are not included in the IUCN Red List of Threatened Species, and the sampling site is not located in any protected area. The field study and laboratory research were conducted by the guidelines provided by Zhengzhou University.
Authors’ contributions
Y.H. analyzed data, prepared the illustration and wrote the draft; Y.Z. analyzed data and prepared the illustration; J.Y. designed the study, prepared the illustrations and revised the manuscript; X.W. designed the study and revised the manuscript. All the authors read and approved the manuscript.
Supplemental Material
Download JPEG Image (417.7 KB)Supplemental Material
Download JPEG Image (1.2 MB)Acknowledgments
We thank Dr. Pengcheng Fu of Luoyang Normal University for offering the leave material and revising the manuscript.
Disclosure statement
All the authors declare no conflicts in this study. The collection of the plant material in this study was carried out in accordance with the national and local regulations.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. OQ446461. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA935068, SAMN33296155, and SRR23490124, respectively.
Additional information
Funding
References
- Bi H, Yang Y, Hu Q, Ma Y. 2020. Characterization of the complete chloroplast genome of Swertia souliei (Gentianaceae). Mitochondrial DNA Part B. 5(2):1901–1902. doi: 10.1080/23802359.2020.1754944.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Brahmachari G, Mondal S, Gangopadhyay A, Gorai D, Mukhopadhyay B, Saha S, Brahmachari AK. 2004. Swertia (Gentianaceae): chemical and pharmacological aspects. Chem Biodivers. 1(11):1627–1651.,. doi: 10.1002/cbdv.200490123.
- Cao Q, Gao Q, Ma X, Zhang F, Xing R, Chi X, Chen S. 2022. Plastome structure, phylogenomics and evolution of plastid genes in Swertia (Gentianaceae) in the Qing-Tibetan Plateau. BMC Plant Biol. 22(1):195. doi: 10.1186/s12870-022-03577-x.
- Chen CL, Ruhfel BR, Li JL, Wang ZF, Zhang LS, Zhang L, et al. 2023. Phylotranscriptomics of Swertiinae (Gentianaceae) reveals that key floral traits are not phylogenetically correlated. J. Integr. Plant Biol. 65:1490–1504. doi: 10.1111/jipb.13464.
- Feng Z, Zheng Y, Jiang Y, Miao Y, Luo G, Huang L. 2022. Complete chloroplast genome of Gentianopsis barbata and comparative analysis with related species from Gentianaceae. Genome. 65(7):363–375. doi: 10.1139/gen-2021-0080.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–W279. doi: 10.1093/nar/gkh458.
- Fu P-C, Sun S-S, Twyford AD, Li B-B, Zhou R-Q, Chen S-L, Gao Q-B, Favre A. 2021. Lineage‐specific plastid degradation in subtribe Gentianinae (Gentianaceae). Ecol Evol. 11(7):3286–3299. doi: 10.1002/ece3.7281.
- Fu PC, Chen SL, Sun SS, Favre A. 2022. Strong plastid degradation is consistent within section Chondrophyllae, the most speciose lineage of Gentiana. Ecol Evol. 12(8):e9205. doi: 10.1002/ece3.9205.
- Ho TN, Liu SW. 2015. A worldwide monograph of Swertia and its allies. Beijing: Science Press.
- Huang L, Chen C, Yang Q, Yang W, Hu Q. 2019. The complete chloroplast genome of Swertia cordata. Mitochondrial DNA Part B Resour. 4(2):3818–3819. doi: 10.1080/23802359.2019.1681310.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z.,. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Ni Y, Li J, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. dx. doi: 10.17504/protocols.io.4r3l27jkxg1y/v1.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Sun S-S, Fu P-C, Zhou X-J, Cheng Y-W, Zhang F-Q, Chen S-L, Gao Q-B.,. 2018. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): Insights into plastid gene loss and molecular evolution. Front Plant Sci. 9:493. doi: 10.3389/fpls.2018.00493.
- Sun SS, Fu PC. 2019. Study on taxonomy and evolution of Gentianeae (Genti anaceae). Acta Bot Sin. 39:363–370.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Xi HC, Sun Y, Xue CY. 2014. Molecular phylogeny of Swertiinae (Gentianaceae Gentianeae) based on sequence data of ITS and matK. Plant Divers. 36:145–156.
- Xiang B, Li X, Qian J, Wang L, Ma L, Tian X, Wang Y. 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules. 21(8):1029. doi: 10.3390/molecules21081029.
- Xu X, Li J, Chu R, Luan M, Wang H, Song K, Wei S, Shi Y, Zhu S, Wei Z, et al. 2021. Comparative and phylogenetic analyses of Swertia L.(Gentianaceae) medicinal plants (from Qinghai, China) based on complete chloroplast genomes. Genet Mol Biol. 45(1):e20210092. doi: 10.1590/1678-4685-GMB-2021-0092.
- Yang Z, Yan J, Zhu G. 2020a. The complete chloroplast genome sequence of Swertia diluta (Gentianaceae). Mitochondrial DNA Part B Resour. 5(3):2763–2764. doi: 10.1080/23802359.2020.1788447.
- Yang L, Xiong F, Xiao Y, Li J, Chen C, Zhou G. 2020b. The complete chloroplast genome of Swertia tetraptera and phylogenetic analysis. Mitochondrial DNA Part B Resour. 5(1):164–165. doi: 10.1080/23802359.2019.1698368.
- Zhang X, Sun Y, Landis JB, Lv Z, Shen J, Zhang H, Lin N, Li L, Sun J, Deng T, et al. 2020a. Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol. 20(1):340. doi: 10.1186/s12870-020-02518-w.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020b. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.

