Abstract
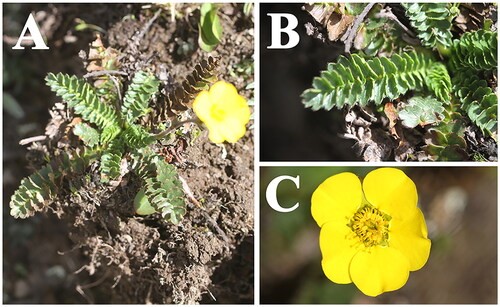
Geum longifolium (Maxim.) Smedmark 2006 belongs to the family Rosaceae, subfamily Rosoideae, tribe Colurieae. Geum longifolium is endemic to China and its whole herb is used in Chinese medicine. Here, the first complete chloroplast (cp) genome of G. longifolium was assembled and annotated based on genome skimming, and its phylogenetic position was investigated using phylogenomic evidence. The cp genome size of G. longifolium was 155,884 bp with the total GC content of 36.7%. Its cp genome presented a typical tetrad structure, composed of a large single copy (LSC) region (85,338 bp), a small single copy (SSC) region (18,358 bp), and a pair of inverted repeat (IR) regions (26,094 bp). The cp genome encoded 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis indicated that G. longifolium was sister to G. elatum Wall. ex G.Don 1832 in current taxa sampling. This study can enrich the chloroplast genomic resource of Geum and lay the foundation for future phylogenetic studies on Geum.
Introduction
Geum longifolium (Maxim.) Smedmark Citation2006 (synonym: Coluria longifolia Maxim. Citation1882; ) is a member of the family Rosaceae Juss., subfamily Rosoideae (Juss.) Arn., tribe Colurieae Rydb. (Smedmark Citation2006). Geum longifolium is endemic to China and distributed in the alpine meadows of Gansu, Qinghai, Sichuan, Xizang, and Yunnan (Li et al. Citation2003). Whole herb of G. longifolium is used as medicine with the effect of hemostasis, pain relief, and heat-clearing (Yü and Kuan Citation1985). The chloroplast (cp) genome of G. longifolium has not been reported to date and its phylogenetic position has not been investigated using the phylogenomic evidence. In the present study, we reported the complete cp genome of G. longifolium for the first time and inferred its phylogenetic relationships with related Geum species. Our study can make a great contribution to further studies on the taxonomy, phylogeny, and population genetics of Geum species.
Materials
Leaf sample of G. longifolium was collected from Qilian County, Qinghai Province, China (38°01.395' N, 100°14.278' E). The specimen was deposited at the herbarium of Inner Mongolia Normal University (NMTC) (http://bio.imnu.edu.cn, Qin-Qin Li, [email protected]) under the voucher number Li QQ BDB1.
Methods
Plant total DNA was extracted from silica gel-dried leaf by modified Cetyl Trimethyl Ammonium Bromide (CTAB) method (Doyle and Doyle Citation1987). Subsequently, the prepared DNA library with an insert size of 300 bp fragments was sequenced by the Illumina NovaSeq 6000 platform in Novogene (Beijing, China). Trimmomatic version 0.33 (Bolger et al. Citation2014) was used to remove adapters after sequencing and a total of 40,106,684-bp raw reads were obtained. The raw reads were assembled by NOVOPlasty version 3.8.3 (Dierckxsens et al. Citation2017), with the cp genome of Geum macrophyllum Willd.1809 (GenBank accession number MT774132; Li and Wen Citation2021) as the reference sequence and its ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcL) gene as the seed and 3,655,282-bp reads were mapped to G. macrophyllum cp genome. Sequencing depth and coverage map of G. longifolium was generated following the protocol of Ni et al. (Citation2023). The cp genome annotation of G. longifolium was conducted using transferring annotations by Geneious prime (Kearse et al. Citation2012), with the cp genome of G. macrophyllum (MT774132) as the reference. Chloroplast Genome Viewer (CPGView) was used to draw the circular cp genome map of G. longifolium and the structure of the genes that are difficult to annotate in the cp genome (Liu et al. Citation2023).
To infer the phylogenetic position of G. longifolium, we conducted a phylogenetic analysis of G. longifolium and its related species. Sixteen cp genome sequences were downloaded from GenBank, including 12 Colurieae accessions and four other Rosoideae species. Based on a previous study (Zhang et al. Citation2017), we selected four Rosoideae species (Agrimonia nipponica Koidz.1930, Potentilla suavis Soják 2008, Rosa multiflora Thunb 1784, and Rubus niveus Thunb.1813) as outgroups. The cp genome sequences of the above 17 accessions were aligned using MAFFT version 7.450 (Katoh and Standley Citation2013) under “auto”settings. Software trimAL version 1.4 (Capella-Gutiérrez et al. Citation2009) was then used to trim the alignment properly with a 0.9 gap threshold. Maximum likelihood (ML) method and Bayesian inference (BI) were used to establish phylogenetic trees respectively. The ML analysis was performed using RAxML version 8 (Stamatakis Citation2014) following Zhang et al. (Citation2020). The BI analysis was conducted using MrBayes ver. 3.2.2 (Ronquist et al. Citation2012) following Tian et al. (Citation2020) and the GTR + I + G model was selected as the best-fit model by PartitionFinder2 (Lanfear et al. Citation2017).
Results
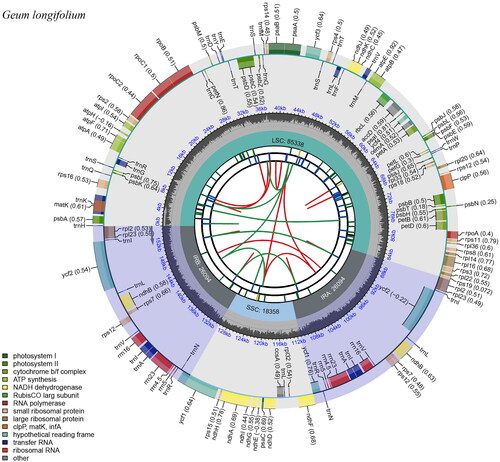
The cp genome of G. longifolium was a circular DNA molecule (GenBank accession number OP161499; ). The cp genome size of G. longifolium was 155,884 bp, with an average depth of 3697.14 × (Figure S1), and the total GC content was 36.7%. Its cp genome presented a typical tetrad structure, consisting of a large single copy (LSC) region (85,338 bp), a small single copy (SSC) region (18,358 bp), and a pair of inverted repeat (IR) regions (26,094 bp). The GC content of LSC, SSC, and IR were 34.4%, 30.9%, and 42.6%, respectively. The cp genome contained 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. In addition, the cp genome contained one pseudogene Ψycf1 located in the IRb/SSC junction. Ten unique genes (clpP, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, ycf3) were cis-splicing and rps12 was trans-splicing (Figure S2).
Figure 2. Genome map of G. longifolium chloroplast genome drawn by Chloroplast Genome Viewer (CPGView, http://www.1kmpg.cn/cpgview). The genome map includes six tracks. From the inward to outward, the first track shows the dispersed repeats which consist of direct repeats and palindromic repeats, connected with red and green arcs. The second track shows the long tandem repeats (blue bars). The third track shows the short tandem repeats or microsatellite sequences as short bars. The fourth track shows the large single copy (LSC), the small single copy (SSC), and inverted repeat (IRa and IRb) regions. The fifth track shows the GC contents along the chloroplast genome. The outermost track shows the genes which are color-coded based on their functional classification. The inner genes are transcribed clockwise, and the outer genes are transcribed anticlockwise.
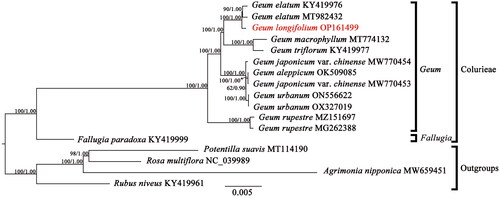
Tree topologies inferred by ML and BI analyses were identical, so the ML tree with bootstrap support values (BS) and Bayesian posterior probabilities (PP) was shown in . The genus Geum was recovered as a monophyletic group in the phylogenetic tree (BS = 100%, PP = 1.00). Within Geum, G. rupestre (Yü et Li) Smedmark Citation2006 is sister to a clade comprising seven other species (G. urbanum L. 1753, G. japonicum var. chinense F.Bolle 1931, G. aleppicum Jacq. 1781, G. triflorum Pursh 1814, G. macrophyllum, G. longifolium and G. elatum Wall. ex G.Don 1832) in current taxa sampling.
Figure 3. The Maximum-likelihood (ML) phylogenetic tree reconstructed based on 13 cp genome sequences from Colurieae plus four other Rosoideae species as outgroups. Values along branch represent ML bootstrap percentages, and Bayesian posterior probabilities respectively. The following sequences were used: Geum elatum KY419976 (Zhang et al. Citation2017), Geum elatum MT982432, Geum longifolium OP161499, Geum macrophyllum MT774132 (Li and Wen Citation2021), Geum triflorum KY419977 (Zhang et al. Citation2017), Geum japonicum var. chinense MW770454, Geum japonicum var. chinense MW770453, Geum aleppicum OK509085 (Zhang et al. Citation2022), Geum urbanum ON556622, Geum urbanum OX327019, Geum rupestre MZ151697, Geum rupestre MG262388 (Duan et al. Citation2018), fallugia paradoxa KY419999 (Zhang et al. Citation2017), Potentilla suavis MT114190 (Li et al. Citation2020), Rosa multiflora NC_039989, Agrimonia nipponica MW659451, and Rubus niveus KY419961 (Zhang et al. Citation2017).
Discussion and conclusion
In this study, using the related bioinformatics methods, the first complete cp genome of G. longifolium was assembled and annotated based on genome skimming. The cp genome of G. longifolium has similar structure and gene size, and consistent gene composition and gene order to that of other Geum species (Duan et al. Citation2018, Li and Wen Citation2021, Zhang et al. Citation2022). Species Geum longifolium was first published under the name of Coluria longifolia Maxim. (1882: 466) and the name was adopted in the Flora of China by Li et al. (Citation2003). Smedmark (Citation2006) made a recircumscription of Geum based on phylogenetic studies of Colurieae (Smedmark and Eriksson Citation2002, Smedmark et al. Citation2003), in which Coluria longifolia were included in Geum as G. longifolium (Maxim.) Smedmark. Our phylogenetic analysis showed that G. longifolium was nested within the Geum species, which supported Smedmark’s taxonomic treatment to place this species within Geum (Smedmark Citation2006). Phylogenetic analysis indicated that G. longifolium was sister to G. elatum in current taxa sampling. This study can enrich the chloroplast genomic resource of Geum and lay the foundation for future phylogenetic studies on Geum.
Ethical approval
No ethical approval is required. Geum longifolium is not an endangered or protected plant.
Authors’ contributions
Conception and design, investigation: Qin-Qin Li, Khasbagan, Soyolt; formal analysis: Jia-Jie Guo, Zhi-Ping Zhang, Qin-Qin Li; the drafting of the paper: Jia-Jie Guo; review and editing of the paper: Zhi-Ping Zhang, Khasbagan, Soyolt, Qin-Qin Li. All authors approved the final version of the paper and agreed to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (2.8 MB)Supplemental Material
Download JPEG Image (1.8 MB)Supplemental Material
Download MS Word (1.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OP161499. The associated BioProject, SRA, and Bio-Sample accession numbers are PRJNA866517, SRR21050029, and SAMN30167531, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Duan N, Liu S, Liu BB. 2018. Complete chloroplast genome of Taihangia rupestris var. rupestris (Rosaceae), a rare cliff flower endemic to China. Conserv Genet Resour. 10(4):809–811. doi: 10.1007/s12686-017-0936-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Li CL, Ikeda H, Ohba H. 2003. Coluria R.Br. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; Vol. 9; p. 289–290.
- Li QQ, Wen J. 2021. The complete chloroplast genome of Geum macrophyllum (Rosaceae: Colurieae). Mitochondrial DNA Part B. 6(2):297–298. doi: 10.1080/23802359.2020.1861562.
- Li QQ, Zhang ZP, Khasbagan. 2020. The complete chloroplast genome of Potentila suavis (Rosaceae: Potentilleae). J Inner Mongolia Norm Univ (Nat Sci Edn). 49(6):471–474. (In Chinese)
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Maximowicz CJ. 1882. Diagnoses plantarum novarum asiaticarum. IV. Bull Acad Imp Sci Saint-Pétersbourg. 27:425–560.
- Ni Y, Li J, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. protocols.io. doi: 10.17504/protocols.io.4r3l27jkxg1y/v1.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Smedmark JEE, Eriksson T. 2002. Phylogenetic relationships of Geum (Rosaceae) and relatives inferred from the nrITS and trnL-trnF regions. Syst Bot. 27(2):303–317.
- Smedmark JEE, Eriksson T, Evans RC, Campbell CS. 2003. Ancient allopolyploid speciation in Geinae (Rosaceae): evidence from nuclear granule-bound starch synthase (GBSSI) gene sequences. Syst Biol. 52(3):374–385. doi: 10.1080/10635150309332.
- Smedmark JEE. 2006. Recircumscription of Geum (Colurieae: Rosaceae). Bot Jahrb Syst. 126(4):409–417. doi: 10.1127/0006-8152/2006/0126-0409.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Tian WJ, Khasbagan, Li QQ. 2020. The complete chloroplast genome of Sibbaldianthe adpressa (Rosaceae: Potentilleae). Mitochondrial DNA Part B. 5(2):1563–1564. doi: 10.1080/23802359.2020.1742601.
- Yü TT, Kuan KC. 1985. Coluria R.Br. In: Yü TT, editor. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; Vol.37; p. 229–232. (In Chinese)
- Zhang PP, Wang L, Lu X. 2022. Complete chloroplast genome of Geum aleppicum (Rosaceae). Mitochondrial DNA Part B. 7(1):234–235. doi: 10.1080/23802359.2021.2024461.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214(3):1355–1367. doi: 10.1111/nph.14461.
- Zhang XH, Khasbagan, Li, QQ 2020. The complete chloroplast genome of Sibbaldia aphanopetala (Rosaceae: Potentilleae). Mitochondrial DNA Part B. 5(3):2026–2027. doi: 10.1080/23802359.2020.1756945.