Abstract
Torenia violacea (Azaola) Pennell 1943 as a traditional Chinese medicine plant, has been used to treat multiple diseases. In this study, we sequenced, assembled, and characterized the complete plastome of T. violacea. The plastome (OQ167784) exhibited a typical quadripartite structure, with a total length of 154,007 bp, comprising a pair of inverted repeats (IRs; 24,809 bp) separated by a large single-copy (LSC) region (85,559 bp) and a small single-copy (SSC) region (18,830 bp). Through genome annotation, we identified 133 genes, including 87 protein-coding genes, eight rRNA genes, and 38 tRNA genes. Phylogenetic analysis revealed a close relationship between T. violacea and T. fournieri. The research results provide basic genetic resources for the development of species identification and investigation of phylogenetic relationships in the Torenia genus.
Introduction
Torenia violacea (Azaola) Pennell 1943, is an annual herbaceous plant belonging to the Torenia genus of the Linderniaceae family, and widely distributed in China. It has a high ornamental value due to its showy purple-red flowers (). The whole plant was also used as a traditional Chinese medicinal herb for clearing heat, expelling dampness, relieving coughs, and other pharmacological effects. Morphologically, T. violacea is very similar to other species of the genus Torenia and can only be distinguished by the detailed characteristics of the flowers (China Citation2013). Before flowering, morphology of the leaves is insufficient to distinguish these species. Therefore, molecular data are required for species delimitation. With the rapid development of high-throughput sequencing technology, the whole chloroplast genome sequence has been widely used for phylogenetic and species delimitation studies (Li et al. Citation2022). To date, there have not been any laboratories using the whole plastome of T. violacea for research for molecular biology species identification. Consequently, we employed next-generation sequencing technology and phylogenetic analysis in this study to publish the whole chloroplast genome of T. violacea and enhance the species delimitation in Torenia.
Figure 1. Photograph of Torenia violacea (the photograph was taken by prof. Xiaoyan Xiang in Mt. Huangshan, China). The foliage of T. violacea exhibits ovate or narrowly ovate shapes, characterized by villous, shallowly serrate margin, acuminate apex. The flowers are in terminal fascicles or solitary in leaf axils, rarely in racemes. Furthermore, the calyx of T. violacea displays purple-red and the corollas pale yellow or white.

Materials and methods
Fresh leaves of T. violacea were collected from Tangkou, Huangshan, Anhui Province, China (30.024°N, 118.188°E) and dried with silica gel. The voucher specimens were deposited in the Herbarium of Anqing Normal University (voucher number: ZLP092302; contact person: Xiaoyan Xiang; email: [email protected]). Total DNA was extracted from dried leaves using the plant genomic DNA kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China).
The DNA library construction and 150-bp paired-end sequencing were performed using the Illumina NovaSeq 6000 platform (Novogene, Tianjin, China), yielding approximately 2 GB of data. Checking and improving the integrality and quality of the whole plastome sequences with Torenia fordii (NCBI accession number: MW309811) as reference plastome, through reference guided mapping using BWA (v0.7.17), SAMtools (v1.13), and IGV (2.15.2), obtained a more accurate plastome genome. The minimum and average read mapping depths for assembled genomes were ×584 and ×4704 (Figure S1), respectively.
The chloroplast genome was assembled using the GetOrganelle pipeline (Jin et al. Citation2020) and annotation was conducted with Plastid Genome Annotator (Qu et al. Citation2019). The annotated chloroplast genome sequence of T. violacea has been deposited in GenBank of the National Center for Biotechnology Information (NCBI accession number: OQ167784). The plastome map of T. violacea was drawn through CPGview (Liu et al. Citation2023). The phylogenetic relationship of T. violacea was reconstructed using the maximum-likelihood (ML) method based on the multiple alignment of Torenia species and seven previously reported chloroplast genomes of Lamiales, with Capsicum annuum (MH559327) and Solanum macrocarpon (MH283714) as the outgroup. All sequences were aligned with MAFFT v.7 (Katoh and Standley Citation2013) using the default settings. ML analysis was conducted using RAxML-HPC v.8.2.8 (Stamatakis Citation2014) with 1000 bootstrap replicates on the CIPRES Science Gateway (https://www.phylo.org/), and the (GTR) + G + I model being used in ML analyses.
Results
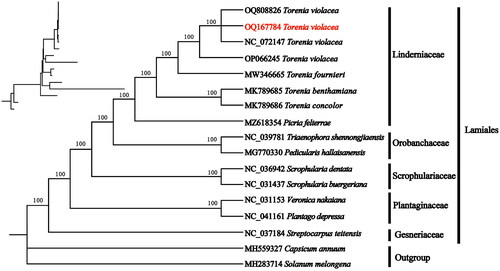
The plastome of T. violacea was 154,007 bp and shared the common quadripartite structure of most angiosperms (). The genome comprises two copies of inverted repeat (IR) (24,809 bp each) separated by a large single-copy (LSC) region (85,559 bp) and a small single-copy (SSC) region (18,830 bp). A total of 133 genes were identified in the cp genome of T. violacea, containing 87 protein-coding genes (PCGs), eight ribosomal RNA genes (rRNAs), and 38 transfer RNA genes (tRNAs). Of these, 11 genes (rsp16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, rp12, ndhB, and ndhA) are cis-spliced, and rps12 is also a trans-spliced gene (Figure S2). The results of phylogenetic research based on ML indicate that T. violacea belonged to the genus Torenia in the Linderniaceae family. The T. violacea and T. fournieri were sister to each other ().
Figure 2. Schematic map of overall features of the chloroplast genome of Torenia violacea. The boxes of different sizes and colors on the outermost circle represent genes and their lengths. The inner and outer boxes of the outermost circle represent genes transcribed clockwise and counter-clockwise. The gray area in the middle circle represents the changes in GC content at different positions, and the regions and lengths represented by the tetrad structure (LSC, SSC, IRa, and IRb) are drawn in different colors on the inner circle.
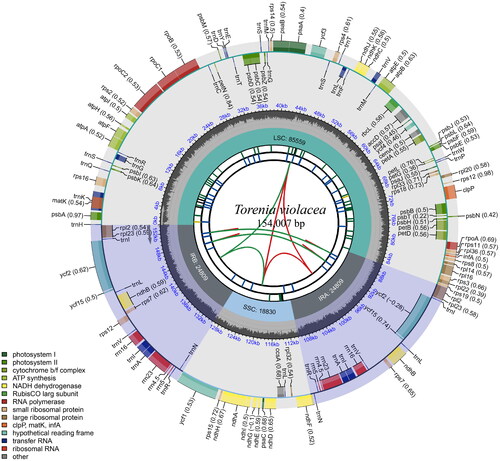
Figure 3. Phylogenetic relationships of Torenia violacea and other species of Lamiales inferred from ML analyses based on the whole chloroplast genome, with Solanum macrocarpon and Capsicum annuum as the outgroup. The numbers on each branches indicate the boot support value of the ML analyses. The sequences used for tree construction are as follows: T. violacea (OQ808826, N_C_072147, OP066245), T. fournieri (MW346665), T. concolor (MK789685, Cheng, Li, et al. Citation2019), T. benthamiana (MK789686, Cheng, Liu, et al. Citation2019), Picria felterrae (MZ618354), Pedicularis hallaisanensis (MG770330, Cho et al. Citation2018), Triaenophora shennongjiaensis (NC_039781, Xia and Wen Citation2018), Scrophularia buergeriana (NC_031437, Yi and Kim Citation2016), Scrophularia dentata (NC_036942), Veronica nakaiana (NC_031437, Choi et al. Citation2016), Plantago depressa (NC_041161), Streptocarpus teitensis (NC_037184, Zhao et al. Citation2019), C. annuum (MH559327, D'Agostino et al. Citation2018), and S. macrocarpon (MH283714, Aubriot et al. Citation2018).
Discussion and conclusions
In this study, the complete plastome of T. violacea was sequenced for the first time, exhibiting a total length of 154,007 bp. A total of 133 genes were annotated, comprising 87 PCGs, eight rRNA genes, and 38 tRNA genes. There were no significant differences in genome size and gene content between T. violacea and other chloroplast genomes in Torenia (Cheng, Li, et al. Citation2019; Cheng, Liu, et al. Citation2019). Phylogenetic analysis indicates a sister relationship between T. violacea and T. fournieri. This suggests a close genetic relationship between the two Torenia genus plants. In addition, species from different families cluster into different monophyletic groups with a 100% bootstrap value. This indicates that chloroplast genomic data can construct robust phylogenetic relationships. In conclusion, our research results provide basic genetic resources for the development of species identification and the investigation of phylogenetic relationships in the Torenia genus.
Ethical approval
The sampling site is not located in any protected area. The research was conducted with the permission of Anqing Normal University of China.
Author contributions
X.Y. Xiang planned and designed the experiments, and drafted the paper; W.H. Zhu and H.Q. Cui performed the experiments and analyzed the data; all authors revised the final version of the paper and have reviewed and approved the manuscript.
Supplemental Material
Download MS Word (327.2 KB)Acknowledgements
We sincerely thank Wei Zhang for submitting the raw data and the chloroplast genome to the GenBank database.
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ167784. The associated BioProject, SequenceRead Archive, and BioSample numbers are PRJNA918330, SRR22967894, and SAMN32573999.
Additional information
Funding
References
- Aubriot X, Knapp S, Syfert MM, Poczai P, Buerki S. 2018. Shedding new light on the origin and spread of the brinjal eggplant (Solanum melongena L.) and its wild relatives. Am J Bot. 105(7):1175–1187. doi: 10.1002/ajb2.1133.
- Cheng XL, Li HL, Wang HX, Nizamani MM, Chen Y. 2019. Complete plastome sequence of Torenia concolor Lindley (Linderniaceae): an ornamental herb. Mitochondrial DNA B Resour. 4(2):2312–2313. doi: 10.1080/23802359.2019.1627949.
- Cheng X-L, Liu Y-M, Wang H-X, Li H-L, Chen Y, Wang L-Y. 2019. Complete plastome sequence of Torenia benthamiana Hance (Linderniaceae), an endemic herb in South China. Mitochondrial DNA B Resour. 4(2):2092–2093. doi: 10.1080/23802359.2019.1618220.
- China ECoFo. 2013. Flora of China. Beijing, China and Missouri: Science Press and Missouri Botanical Garden Press.
- Cho WB, Lee DH, Choi IS, Lee JH. 2018. The complete chloroplast genome of hemi-parasitic Pedicularis hallaisanensis (Orobanchaceae). Mitochondrial DNA B Resour. 3(1):235–236. doi: 10.1080/23802359.2018.1437820.
- Choi KS, Chung MG, Park S. 2016. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Front Plant Sci. 7:355. doi: 10.3389/fpls.2016.00355.
- D'Agostino N, Tamburino R, Cantarella C, De Carluccio V, Sannino L, Cozzolino S, Cardi T, Scotti N. 2018. The complete plastome sequences of eleven Capsicum genotypes: insights into DNA variation and molecular evolution. Genes. 9(10):503. doi: 10.3390/genes9100503.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, Huang Y, Zhou Q, Zhu Q. 2022. Complete chloroplast genome sequences of three Aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 23(1):218–234. doi: 10.1186/s12864-022-08400-3.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Xia Z, Wen J. 2018. The complete chloroplast genome of the endangered species Triaenophora shennongjiaensis (Orobanchaceae s.l.). Mitochondrial DNA B Resour. 3(2):506–507. doi: 10.1080/23802359.2018.1467242.
- Yi DK, Kim KJ. 2016. The two complete plastomes from Scrophularia (Scrophulariaceae): Scrophularia buergeriana and S. takesimensis. Mitochondrial DNA B Resour. 1(1):710–712. doi: 10.1080/23802359.2016.1225528.
- Zhao X, Yan M, Ding Y, Huo Y, Yuan Z. 2019. Characterization and comparative analysis of the complete chloroplast genome sequence from Prunus avium 'Summit'. PeerJ. 7:e8210. doi: 10.7717/peerj.8210.
