Abstract
Swertia japonica (Schult.) Makino is a traditional medicinal plant in Japan for which the chloroplast genome has not been previously reported. The complete chloroplast genome of S. japonica was determined using a high-throughput sequencing technique. The total length of the S. japonica chloroplast genome was 153,208 bp, and comprised a large single-copy region of 83,319 bp, and a small single-copy region of 18,375 bp, separated by a pair of 25,757 bp inverted repeat regions. A phylogenetic analysis, based on the obtained chloroplast genome, indicated that S. japonica is closely related to S. diluta, S. franchetiana, S. kouitchensis, S. mussotii, and S. punicea. The presented chloroplast genome will be useful for further taxonomic, pharmacological and evolutionary studies of Swertia.
Introduction
Swertia japonica (Schult.) Makino (Makino Citation1910) is a biennial plant that is found across Japan and Korea. This species is a part of traditional Japanese medicine, and is commonly used for gastrointestinal diseases including nausea, gastroparesis, and gastric atony (Kimura and Sumiyoshi Citation2011). Moreover, the plant has potential for antipyretic, anthelmintic, tonic, anti-periodic, cathartic, asthma, leucorrhoea, analeptic, anti-inflammatory, and relaxing the pregnant uterus (Rai et al. Citation2016). The genus includes other species that are used for medicinal purposes, but the taxonomy of the genus remains unresolved due to polyphyly (Favre et al. Citation2010). Therefore, insights into the genomic features and phylogenetic relationships among Swertia species will prove valuable to the taxonomic revision of this genus. In this study, we report the complete chloroplast genome of S. japonica based on Illumina paired-end sequencing data. In addition, phylogenetic relationships of the genus Swertia and related genera were reconstructed by utilizing published sequences of related species.
Materials and methods
Sample leaves were collected from Tsugeno in Aichi Prefecture, Japan (34.8639N, 137.5775E) on 16 October 2022 (GenBank BioSample, SAMD00578002). The sample was identified and collected by the author, and the voucher specimen was deposited in the Herbarium of the National Museum of Nature and Science, Japan (https://tbg.kahaku.go.jp/english/index.php, Atsushi Ebihara, [email protected]) under the accession number TNS1347890 ().
Figure 1. The analyzed sample of Swertia japonica. The photograph was taken by Watanabe Yoichi at Tsugeno, Aichi Prefecture, Japan, on 16 October 2022. Swertia japonica is a biennial plant 5–20 cm in height, with a corolla diameter of 2–3 cm and petal with nectaries.

Total genomic DNA was extracted from dried leaves using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany). The extracted DNA was used to prepare a sequencing library using the TruSeq DNA PCR-Free Kit (Illumina, San Diego, CA) and then sequenced using 150 base-length read chemistry in a paired-end flow cell on the NovaSeq 6000 platform (Illumina, San Diego, CA); the library preparation and sequencing were conducted by Macrogen (Seoul, South Korea). The 4.8 Gbases of raw data was trimmed by clipping adaptor sequences and removing reads with low quality using fastp v. 0.20.0 (Chen et al. Citation2018). Trimmed reads were deposited in the GenBank (accession number, DRR441012). Trimmed reads were assembled using GetOrganelle v.1.7.5 (Jin et al. Citation2020) with the following settings: -F embplant_pt; and default settings for other options. The assembled genome was annotated based on BLAST searches using GeSeq (Tillich et al. Citation2017) with default settings. To verify complete coverage of the chloroplast genome, the trimmed reads were mapped to the assembled genome using BWA (Li and Durbin Citation2009) with default settings. To facilitate the mapping to the circular genome, the sequence was linearized and duplicated 700 bp from one end to the opposite end. The genome map and cis/trans-splicing genes maps were drawn using CPGView (Liu et al. Citation2023). The annotated chloroplast genome was deposited in the GenBank (accession number, LC744566). To investigate the phylogenetic position of S. japonica, chloroplast genome sequences of related species were obtained from the GenBank database. A total of 23 Swertia species based on the research by Yang et al. (Citation2022), and three Gentiana species, which served as outgroups of Swertia, were used. A total of 69 genes longer than 150 bp, which were shared among the species were extracted, and each orthologous gene was aligned using MAFFT v.7.505 (Katoh and Standley Citation2013) with default settings, and then these alignments were combined. A phylogenetic tree by the maximum likelihood method was constructed using RAxML v.8.2.0 (Stamatakis Citation2014) under the GTR + G + I substitution model with 1000 bootstrap replicates.
Results
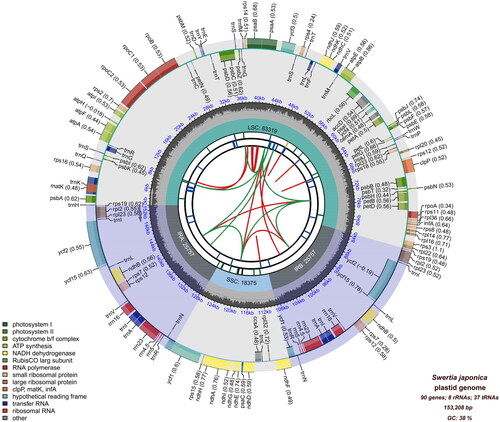
The total length of the complete chloroplast genome of S. japonica was 153,208 bp, with an average coverage of ×1578.9 and GC content of 38% (, Supplemental Figure 1). A large single-copy (LSC: 83,319 bp), a small single-copy (SSC: 18,375 bp), and two inverted repeat (IR: 25,757 bp) regions made up the quadripartite structure of the chloroplast genome. A total of 135 genes were annotated, including 90 protein-coding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes, of which 20 genes were duplicated in the IR regions. Twelve cis-splicing genes and one trans-splicing gene, rps12, were identified ().
Figure 2. The complete chloroplast genome of Swertia japonica. The map contains six tracks. From the center outward, the first track displays the dispersed repeats. The dispersed repeats consist of direct and palindromic repeats, connected with red and green arcs. The second track shows long tandem repeats as short blue bars. The third track shows short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. Optional codon usage bias is displayed in parentheses after the gene name. Genes belonging to different functional groups are color-coded. Genes on the inside and outside of the map are transcribed in clockwise and counterclockwise directions, respectively.
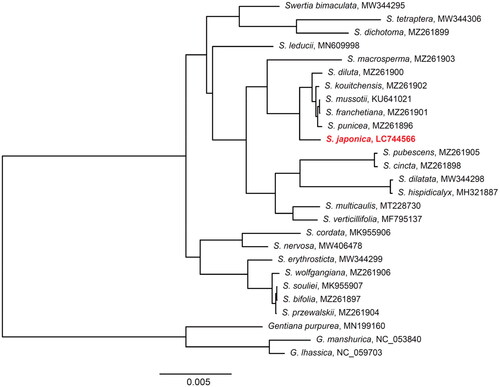
The phylogenetic analysis showed that 24 Swertia species were monophyletic with the highest probability (). Swertia japonica was closest related to the clade consisting S. diluta, S. franchetiana, S. kouitchensis, S. mussotii, and S. punicea ().
Figure 3. A maximum-likelihood (ML) tree of 24 Swertia species and three Gentiana species, which were used as outgroups. Bootstrap probabilities of all nodes are 100%. The genome of the species identified with red-bold font was presented in this study. The following sequences were used: S. bimaculata MW344295 (Xu et al. Citation2022), S. tetraptera MW344306 (Xu et al. Citation2022), S. dichotoma MZ261899 (Cao et al. Citation2022), S. leducii MN609998 (Yang et al. Citation2019), S. macrosperma MZ261903 (Cao et al. Citation2022), S. diluta MZ261900 (Cao et al. Citation2022), S. kouitchensis MZ261902 (Cao et al. Citation2022), S. mussotii KU641021 (Xiang et al. Citation2016), S. franchetiana MZ261901 (Cao et al. Citation2022), S. punicea MZ261896 (Cao et al. Citation2022), S. pubescens MZ261905 (Cao et al. Citation2022), S. cincta MZ261898 (Cao et al. Citation2022), S. dilatata MW344298 (Xu et al. Citation2022), S. hispidicalyx MH321887, S. multicaulis MT228730 (Zhang et al. Citation2020), S. verticillifolia MF795137, S. cordata MK955906 (Huang et al. Citation2019), S. nervosa MW406478, S. erythrosticta MW344299 (Xu et al. Citation2022), S. wolfgangiana MZ261906 (Cao et al. Citation2022), S. souliei MK955907, S. bifolia MZ261897 (Cao et al. Citation2022), S. przewalskii MZ261904 (Cao et al. Citation2022), G. purpurea MN199160, G. manshurica NC_053840 (Liang et al. Citation2020), and G. lhassica NC_059703 (Fu et al. Citation2021).
Discussion and conclusions
The S. japonica chloroplast genome was similar in structure to the chloroplast genome of other Swertia species (Xiang et al. Citation2016; Huang et al. Citation2019; Cao et al. Citation2022; Xu et al. Citation2022; Yang et al. Citation2022). The result of the phylogenetic analysis in this study concur with the topology of Swertia (without the inclusion of S. japonica) presented in the previous study (Yang et al. Citation2022). The phylogenetic analysis suggests that S. japonica is closest related to the clade comprising S. diluta, S. franchetiana, S. kouitchensis, S. mussotii, and S. punicea. Among these relatives, S. japonica, S. mussotii, and S. diluta are prevalent in traditional Chinese and Japanese medicine (Xiang et al. Citation2016).
This study provides a reliable chloroplast genome of S. japonica. The presented complete chloroplast genome can contribute to taxonomic, pharmacological, and evolutionary studies that focus on Swertia and related genera.
Author contributions
W. Y. designed the study, collected the sample, conducted molecular experiments, generated, analyzed, and visualized data, as well as wrote the manuscript.
Ethical approval
The material involved in this article does not involve any ethical conflicts. This species is not endangered according to the CITES catalogue or IUCN Red List, and the sample was not collected from a natural reserve, so the collection did not require any specific permissions or licenses.
Supplemental Material
Download MS Word (1,007.3 KB)Disclosure statement
The author declares no conflicts of interest.
Data availability statement
The data that support the findings presented in this study are openly available in GenBank (NCBI, https://www.ncbi.nlm.nih.gov) under the reference number LC744566. The associated BioProject, SRA, and Bio-Sample numbers are PRJDB15262, DRR441012, and SAMD00578002, respectively.
Additional information
Funding
References
- Cao Q, Gao Q, Ma X, Zhang F, Xing R, Chi X, Chen S. 2022. Plastome structure, phylogenomics and evolution of plastid genes in Swertia (Gentianaceae) in the Qing‑Tibetan Plateau. BMC Plant Biol. 22(1):195. doi: 10.1186/s12870-022-03577-x.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi: 10.1093/bioinformatics/bty560.
- Favre A, Yuan YM, Küpfer P, Alvarez N. 2010. Phylogeny of subtribe Gentianinae (Gentianaceae): biogeographic inferences despite limitations in temporal calibration points. Taxon. 59(6):1701–1711. doi: 10.1002/tax.596005.
- Fu PC, Twyford AD, Sun SS, Wang HY, Xia MZ, Tan CX, Zhou XJ, Chen SL. 2021. Recurrent hybridization underlies the evolution of novelty in Gentiana (Gentianaceae) in the Qinghai-Tibetan Plateau. AoB Plants. 13(1):plaa068. doi: 10.1093/aobpla/plaa068.
- Huang L, Chen C, Yang Q, Yang W, Hu Q. 2019. The complete chloroplast genome of Swertia cordata. Mitochondrial DNA B Resour. 4(2):3818–3819. doi: 10.1080/23802359.2019.1681310.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kimura Y, Sumiyoshi M. 2011. Effects of Swertia japonica extract and its main compound swertiamarin on gastric emptying and gastrointestinal motility in mice. Fitoterapia. 82(6):827–833. doi: 10.1016/j.fitote.2011.04.008.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324.
- Liang Y, Meng X, Mu Z, Qian J, Duan B, Xu L. 2020. The complete chloroplast genome and phylogenetic analysis of Gentiana manshurica Kitag from China. Mitochondrial DNA B Resour. 5(2):1625–1626. doi: 10.1080/23802359.2020.1745107.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Makino T. 1910. Observations on the Flora of Japan. Bot Mag Tokyo. 24:290–307.
- Rai A, Nakamura M, Takahashi H, Suzuki H, Saito K, Yamazaki M. 2016. High-throughput sequencing and de novo transcriptome assembly of Swertia japonica to identify genes involved in the biosynthesis of therapeutic metabolites. Plant Cell Rep. 35(10):2091–2111. doi: 10.1007/s00299-016-2021-z.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Xiang B, Li X, Qian J, Wang L, Ma L, Tian X, Wang Y. 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules. 21(8):1029. doi: 10.3390/molecules21081029.
- Xu X, Li J, Chu R, Luan M, Wang H, Song K, Wei S, Shi Y, Zhu S, Wei Z. 2022. Comparative and phylogenetic analyses of Swertia L. (Gentianaceae) medicinal plants (from Qinghai, China) based on complete chloroplast genomes. Genet Mol Biol. 45:e20210092.
- Yang C-W, Liu Y-Y, Zhang Y-M, Ma X-H, Qian Z-G, Li G-D. 2019. Complete chloroplast genome sequence of Swertia mileensis (Gentianaceae): a medicinal species endemic to China. Mitochondrial DNA B Resour. 4(2):4166–4167. doi: 10.1080/23802359.2019.1693303.
- Yang L, Li J, Zhou G. 2022. Comparative chloroplast genome analyses of 23 species in Swertia L. (Gentianaceae) with implications for its phylogeny. Front Genet. 13:895146. doi: 10.3389/fgene.2022.895146.
- Zhang X, Sun Y, Landis JB, Lv Z, Shen J, Zhang H, Lin N, Li L, Sun J, Deng T, et al. 2020. Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol. 20(1):340. doi: 10.1186/s12870-020-02518-w.
