Abstract
Campsis radicans (L.) Bureau Citation1864, a species of Bignoniaceae, has a widespread paleotropical distribution and is utilized for horticultural and traditional Chinese medicinal purposes. Despite the plant’s significance, its genetic diversity must be better understood. In this study, we have successfully assembled and characterized the complete plastome of C. radicans, marking a significant advancement toward comprehending its genetic composition. The plastome is 153,630 bp long and harbors 130 genes, including 86 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Our phylogenomic analysis of the representative species of Bignoniaceae indicated that C. radicans formed a monophyletic sister clade of Campsis with C. grandiflora. These findings are crucial for conserving and utilizing this important plant species. They also highlight the potential for future research into the evolution and preservation of C. radicans, which could be advantageous in pharmaceutical applications.
Introduction
Campsis, a genus within the Bignoniaceae family, comprises two species of woody vines with a climbing tendency found in North America, China, and Japan. The model species of Campsis is Campsis grandiflora (Thunb.) K. Schum., characterized by features such as aerial roots, odd-pinnate compound leaves, large red or orange-red flowers, and capsules containing numerous seeds with semi-transparent membranous wings. These plants exhibit a climbing growth habit, are typically deciduous, and tend to bloom during the summer. They are well-adapted to harsh climates, thrive in full sunlight, and employ aerial roots for climbing support (Zhang and Thawatchai Citation1998).
Campsis radicans (L.) Bureau (Citation1864), another member of the Campsis genus, is a widely distributed paleotropical species used as a horticultural plant and in traditional Chinese medicine (TCM). Its flowers are employed in TCM under the name Campsis Flos, prescribed for conditions such as irregular menstruation, dysmenorrhea, postpartum breast enlargement, redness of rubella, itchy skin, and acne (Chinese Pharmacopeia, 2020 edition). This beautiful liana is typically cultivated in courtyards to enhance the esthetic appeal of the surrounding environment. Campsis radicans stands out from common fruits and vegetables due to its high concentration of phenolic compounds (Cai et al. Citation2004). These compounds have demonstrated potent antioxidant and anticancer properties. For instance, C. radicans flowers are a rich source of liver-protective compounds, exhibiting similar properties to other hepatoprotective TCMs (He et al. Citation2022). Despite its pharmaceutical potential, the genetic diversity of C. radicans remains relatively unexplored. Therefore, this study aimed to assemble and characterize the plant’s complete plastome, which could serve as a foundation for future research on the evolution, conservation, and potential medicinal applications of C. radicans.
Materials
The fresh leaves used for sequencing were obtained from Peony District, Heze City, Shandong Province, China (35°15′9.27″N, 115°29′44.44″E) (), and a specimen has been deposited in the Heze University Herbarium (contact person: Hongqin Li, [email protected]) under the specimen number HZ20220801. Genomic DNA was extracted using the plant genomic DNA kit (Tiangen Biotech, Beijing, China).
Figure 1. Field photos of Campsis radicans. The author Liqiang Wang shot the photo at the position of 35o 15' 9.27" N, 115o 29' 44.44" E. Main identifying traits: 9 ∼ 11 leaflets, calyx five isolobes, shallow in the division, lobed to about one-third. The corolla is slender funnel-shaped, with distinct brownish-red longitudinal stripes inside, and the tube is three times the size of the calyx.

Methods
The total genomic DNA extracts were fragmented into approximately 300 bp short-insert fragments to generate libraries, which were then sequenced using Illumina NovaSeq 6000 technology platforms at Wuhan Benagen Technology Company Limited in Wuhan, China. Trimmomatic (v0.35) (Bolger et al. Citation2014) was implemented to filter the raw reads by eliminating adapters and low-quality bases. Following the filtering process, roughly 24 GB of clean reads were assembled using GetOrganelle (v1.7.1) (Jin et al. Citation2020). The finished plastome was annotated using CPGAVAS2 (Shi et al. Citation2019) and manually adjusted through Apollo (Pontius Citation2018). Eventually, the annotated plastome was submitted to GenBank using Bankit and assigned the accession number OQ198454. The CPGview tool was utilized to exhibit the circular genome map of the novel plastome.
To establish the phylogenetic relationship of C. radicans, 19 species plastomes of Bignoniaceae were downloaded from GenBank that exhibited the highest degree of similarity to C. radicans based on plastome blast results. In addition, the outgroup is Nymphaea tetragona Georgi. The entire plastome sequences with default parameters were aligned using MAFFT software (https://mafft.cbrc.jp/alignment/software/) (Katoh and Standley Citation2016). Following this, a maximum-likelihood (ML) phylogenetic tree was constructed using IQ-TREE (v2.0) (Nguyen et al. Citation2015) with the Best-fit model of TVM + F+G4 along with 1000 bootstrap replicates. To check the reliability of the tree, the researchers used the Shimodaira-Hasegawa (SH) (Shimodaira and Hasegawa Citation1999) and proximately unbiased (AU) (Shimodaira Citation2002) methods embedded in IQ-TREE. The primary test script used was iqtree -s input_mafft.phy -m TVM + F+G4 -z genome.unconstrain.constrain.treels -zb 10000 -zw -au.
Results and discussion
The C. radicans plastome sequence measures 153,630 bp long and presents a typical quadripartite structure. It consists of two IR regions, each 25,586 bp in length, separated by a large LSC region of 84,608 bp and a small SSC region of 17,850 bp (). Mapping experiments demonstrate the high reliability of the genome assembly (Figure S1). The plastome exhibits a varying GC content distribution, with an overall GC content of 38.2%. The highest GC content is found in the IR regions (43.2%), whereas the corresponding values are 36.2% and 32.8% for the LSC and SSC regions, respectively. Notably, the structure of C. radicans plastome bears similarities to that of C. grandiflora (Chen et al. Citation2022). Upon comparative analysis with the Arabidopsis thaliana plastome, we observed that the LSC of C. radicans underwent rearrangement (Figure S2), a phenomenon also seen in C. grandiflora (Chen et al. Citation2022).
Figure 2. Gene map of the complete chloroplast genome of Campsis radicans. The species name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows dispersed repeats, including direct and palindromic repeats connected by red and green arcs. The second track displays long tandem repeats as blue bars, while the third displays short tandem repeats or microsatellite sequences as differently colored short bars. These colors correspond to the type and description of each repeat, with black representing complex repeats, green for repeat unit size 1, yellow for size 2, purple for size 3, blue for size 4, orange for size 5, and red for size 6. The fourth track displays the SSC, IRa, IRb, and LSC regions. The fifth track shows the GC content along the genome, while the sixth track sounds the genes. The gene names are followed by optional information about codon usage bias and color-coded based on their functional classification. The inner genes are transcribed clockwise, and the outer genes are transcribed anticlockwise. The functional type of the genes is shown in the bottom left corner.
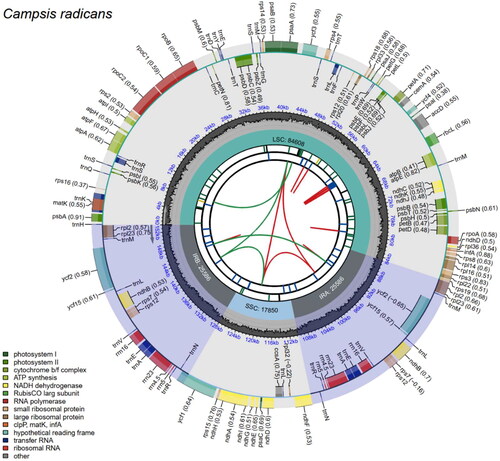
The C. radicans plastome was predicted 130 genes, including 86 protein-coding genes (PCGs), eight rRNA genes, and 36 tRNA genes. Seven unique PCGs (rps12, rps7, rpl2, rpl23, ndhB, ycf15 and ycf2), seven unique tRNA genes (trnA, trnE, trnL, trnM, trnN, trnR and trnV) and four unique rRNA genes (rrn16S, rrn23S, rrn4.5S, rrn5S) were located at the IR regions. In the whole plastome, ten PCGs (rps16, atpF, rpoC1, accD, petD, petB, rpl16, rpl2, ndhB, and ndhA) comprise one intron each, while two PCGs (ycf3, rps12) contain two introns; in addition, five tRNA genes (trnK-UUU, trnS-CGA, trnL-UAA, trnE-UUC, and trnA-UGC) contain one intron. The rps12 gene was subject to trans-splicing. The structures of the cis-splicing and trans-splicing PCG genes are shown in Figure S3. The length of the protein-coding genes, tRNA genes, and rRNA genes in the plastome are 2,7701bp, 3,923bp, and 9,276bp, respectively, making up 18.0%, 2.6%, and 6.0% of the total genome length. Notably, although the size of C. radicans plastome is only 673 bp shorter than that of C. grandiflora, the number of predicted genes in C. radicans plastome was higher than that of C. grandiflora (Chen et al. Citation2022).
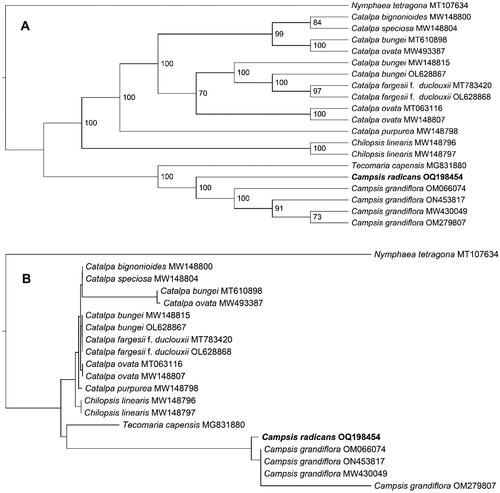
The ML phylogenetic tree demonstrated that the C. radicans and all four C. grandiflora plants formed a monophyletic clade, the sister clade of Tecomaria capensis, with 100% bootstrap values (). This result corresponded with a prior study (Chen et al. Citation2022) indicating a close relationship between Campsis and Tecomaria species. Despite some nodes having comparatively low bootstrap values, the topological structure of the ML tree was significantly included, with a p-SH value of 1.000 and a p-AU value of 0.536, respectively. Consequently, the phylogenetic analysis has provided a dependable account of C. radicans’ evolution.
Figure 3. The Maximum-Likelihood phylogeny of Campsis radicans and its close relatives using whole genome sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree (A). The corresponding phylogram tree is shown in panel B. We downloaded 19 Campsis species plastomes from GenBank, C. grandiflora (Thunb.) K.Schum. (MW430049) (Chen et al. Citation2022), C. grandiflora (OM066074) (Ngai et al. Citation2023), C. grandiflora (OM279807), C. grandiflora (ON453817), Catalpa bignonioides walter (MW148800) (Dong et al. Citation2022), Catalpa bungei C.A.Mey. (MT610898), Catalpa bungei (MW148815) (Dong et al. Citation2022), Catalpa bungei (OL628867) (Li et al. Citation2022), Catalpa fargesii f. duclouxii (dode) Gilmour (MT783420) (Ma et al. Citation2020), Catalpa fargesii f. duclouxii (OL628868) (Li et al. Citation2022), Catalpa ovata G.Don (MT063116) (Wang et al. Citation2020), Catalpa ovata (MW148807) (Dong et al. Citation2022), Catalpa ovata (MW493387), Catalpa purpurea griseb. (MW148798) (Dong et al. Citation2022), Catalpa speciosa teas (MW148804) (Dong et al. Citation2022), chilopsis linearis (cav.) sweet (MW148796) (Dong et al. Citation2022), Catalpa linearis (MW148797) (Dong et al. Citation2022), Tecomaria capensis (Thunb.) spach (MG831880) (Fonseca and Lohmann Citation2018). Nymphaea tetragona Georgi (MT107634) (Sun et al. Citation2021), from the nymphaeaceae, served as the outgroup. The new C. radicans plastome in this study were labeled in bold font.
Conclusions
This study presents the primary characterization of the genome of C. radicans for the first time, which exhibits a typical annular tetrad structure with a size of 153,630 bp and 130 predicted genes. A rearrangement event has occurred in the plastome structure. Based on the phylogenetic analysis, C. radicans and C. grandiflora cluster within Bignoniaceae.
Ethical approval
The C. radicans specimen is not an endangered species. It requires no specific permissions or licenses. Research on this species, including the collection of plant material, was conducted following the guidelines provided by Heze University in this study.
Author contributions
The manuscript includes the contributions of all authors. Liqiang Wang was mainly responsible for the study design. Hongqin Li identified the species and collected samples of the species. Liqiang Wang assembled and annotated the palstome. Hongqin Li and Changhao Ma analyzed the structure of the plastomes and drafted the manuscript. All authors approved the version to be published and agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (136.6 KB)Supplemental Material
Download MS Word (751.5 KB)Supplemental Material
Download MS Word (175.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence of C. radicans in this study has been submitted to the NCBI database under the accession number OQ198454. https://www.ncbi.nlm.nih.gov. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA928567, SAMN32935701, and SRR23250959.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Bureau LÉ. 1864. Campsis radicans (Linnaeus) Seemann ex Bureau. Monogr Bignon. 2(Atlas):16.
- Cai Y, Luo Q, Sun M, Corke H. 2004. Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci. 74(17):2157–2184. doi:10.1016/j.lfs.2003.09.047.
- Chen HM, Chen ZE, Du Q, Jiang M, Wang B, Liu C. 2022. Complete chloroplast genome of Campsis grandiflora (Thunb.) schum and systematic and comparative analysis within the family Bignoniaceae. Mol Biol Rep. 49(4):3085–3098. doi:10.1007/s11033-022-07139-0.
- Dong W, Liu Y, Li E, Xu C, Sun J, Li W, Zhou S, Zhang Z, Suo Z. 2022. Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol Phylogenet Evol. 166:107330. doi:10.1016/j.ympev.2021.107330.
- Fonseca LHM, Lohmann LG. 2018. Combining high-throughput sequencing and targeted loci data to infer the phylogeny of the “Adenocalymma-Neojobertia” clade (Bignonieae, Bignoniaceae). Mol Phylogenet Evol. 123:1–15. doi:10.1016/j.ympev.2018.01.023.
- He S, Yi Y, Hou D, Fu X, Zhang J, Ru X, Xie J, Wang J. 2022. Identification of hepatoprotective traditional Chinese medicines based on the structure-activity relationship, molecular network, and machine learning techniques. Front Pharmacol. 13:969979. doi:10.3389/fphar.2022.969979.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TA-O, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942. doi:10.1093/bioinformatics/btw108.
- Li F, Liu Y, Wang J, Xin P, Zhang J, Zhao K, Zhang M, Yun H, Ma W. 2022. Comparative analysis of chloroplast genome structure and phylogenetic relationships among six taxa within the genus Catalpa (Bignoniaceae). Front Genet. 13:845619. doi:10.3389/fgene.2022.845619.
- Ma WJ, Xu SZ, Yang GJ, Liu Y, Xiang XG, Wang JH. 2020. The complete chloroplast genome sequence of Catalpa fargesii f. duclouxii (Bignoniaceae). Mitochondrial DNA Part B. 5(3):3427–3429. doi:10.1080/23802359.2020.1823267.
- Ngai HL, Kong BLH, Lau DTW, Shaw PC. 2023. Differentiation of Lingxiaohua and Yangjinhua by chloroplast genome sequencing and DNA barcoding markers. Genome. 66(2):21–33. doi:10.1139/gen-2022-0063.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ, Tree IQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Pontius E. 2018. Apollo. Ann Emerg Med. 72(5):616. doi:10.1016/j.annemergmed.2018.06.016.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508. doi:10.1080/10635150290069913.
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of Log-Likelihoods with applications to phylogenetic inference. Mol Biol Evol. 16(8):1114–1116. doi:10.1093/oxfordjournals.molbev.a026201.
- Sun C, Chen F, Teng N, Xu Y, Dai Z. 2021. Comparative analysis of the complete chloroplast genome of seven Nymphaea species. Aquat Bot. 170(1):103353. doi:10.1016/j.aquabot.2021.103353.
- Wang Y, Zhao Y, Wang K, Wang L, Feng Y, Qi L, Luo Y, Ji Y, Gong X. 2020. The complete chloroplast genome of Catalpa ovata (Bignoniaceae): an important ornamental and medicinal plant. Mitochondrial DNA Part B. 5(2):1675–1676. doi:10.1080/23802359.2020.1742226.
- Zhang ZY, Thawatchai S. 1998. Bignoniaceae. Flora of China. 18:213–225.
