Abstract
Scutellaria franchetiana H.Lév. 1911 is an important medicinal plant distributed on hillside wetlands at an altitude of 830-2300 m. The total length of complete chloroplast genome of S. franchetiana 151,852 bp of 38.37% GC content. It is a typical quadratic structure with four subregions consisting of a large single-copy region (83,894 bp), a small single-copy region (17,520 bp), and a pair of inverted repeats (25,219 bp). In the chloroplast genome of S. franchetiana, 130 genes were detected, comprising 86 protein-encoding genes, eight ribosomal RNA (rRNA) genes, and 36 transfer RNA (tRNA) genes. The comparative analysis of complete chloroplast genome sequences including 68 common genes of 27 plants indicates that S. franchetiana has a close relative relationship with Scutellaria orthocalyx and Scutellaria meehanioides. This work reported the first chloroplast genome of S. franchetiana, which provided a potential reference for studying phylogenetic relationships of the Scutellaria genus.
Introduction
Scutellaria franchetiana H.Lév. 1911 belongs to Sect. Maschalostachys of Scutellaria genus and predominantly inhabits hillside wetlands at altitudes ranging from 830 to 2,300 meters. The Scutellaria genus, a member of the Labiatae family, comprises more than 300 species that are globally distributed among angiosperms (Bruno et al. Citation2002). Although the species of the genus Scutellaria are very rich, the Scutellaria plant sequence is still relatively few in NCBI genebank (25 complete chloroplast genomes include S. franchetiana, lastest date:2023.08.24). Therefore, there are many pharmacological studies on Scutellaria but few studies on systematic evolution at present. Scutellaria plants have been used as medicine in China since 2000 years ago, with the functions of clearing away heat and dampness, resisting inflammation and oxidation, and stopping bleeding (Shen et al. Citation2021). However, the research focus is on S. baicalensis, and there are few studies on the genetic relationship and pharmacological activities of other plants in Scutellaria (Islam et al. Citation2022). The chloroplast genome of S. franchetiana has not been published, which leads to its systematic genetic location is still unclear. For this reason, we constructed a high-quality assembled chloroplast to provide a reference to explore phylogenetic relationship of Scutellaria genus.
Materials and methods
We collected the fresh leaves of S. franchetiana from Mountain Jinyun, Chongqing (Geospatial coordinates: N29.83, E106.39). The original plant was identified as S. franchetiana by Professor Weikai Gao ( and Figure S1). We used the genomic DNA kit (Tiangen Biotech, Beijing) to extract the total DNA of fresh leaves. The genome sequence was performed on the Hiseq 2500 platform (Illumina, San Diego, CA, USA). A specimen was deposited at Guizhou Tobacco Company Anshun Tobacco Company (Prof. Weikai Gao, E-mail: [email protected]) under the voucher number 202211212.
Figure 1. S. franchetiana H.Lév. (These photographs were taken by Prof. Weikai Gao). (A) S. franchetiana in its natural habitat (B) The flowers of S. franchetiana.

In total, 5.81 G of raw data were assembled into a chloroplast genome using NOVOPlasty (version 2.7.2) (Dierckxsens et al. Citation2017). Annotation of S. franchetiana sequence was performed using CPGAVAS2 (Shi et al. Citation2019). The average read mapping depths of the assembled genome were 2,113× (Figure S2). By aligning all raw reads against the assembled genome the genome sequence was confirmed using SAMtools v1.9 and BWA v0.7.17 (WGS500 Consortium, 2014) in GeIS (the environment of Genome Information System; https://geis.infoboss.co.kr/). A circular map of the chloroplast genome and a schematic map of the cis- and trans-splicing genes were drawn by CPGView ( and Figure S3) (Liu et al. Citation2023).
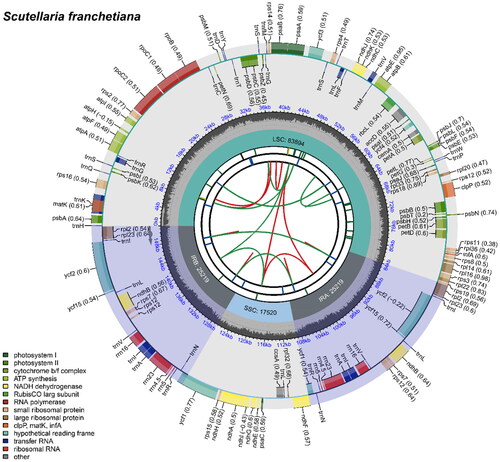
Figure 2. The chloroplast genome map of S. franchetiana. The circle map of chloroplast genome map of S. franchetiana. Genes are depicted within distinctive colored boxes encircling the outer circle, with genes transcribed in a clockwise direction inside the circle and those transcribed counter-clockwise positioned outside. The circles closest to the center are indicated by red and green arcs for forward and reverse repeats, respectively. The inner circle displays a gray region, indicating the GC content, while the quadripartite structure (LSC, SSC, IRA, and IRB) is accurately represented within the inner circle.
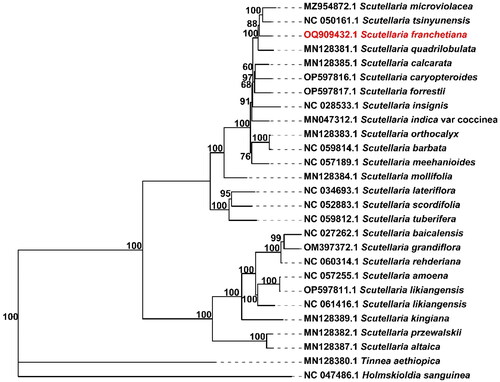
To investigate the phylogenetic relationship of Scutellaria genus, we downloaded chloroplast genomes sequences in NCBI including 24 Scutellaria plants and two outgroup plants (Holmskiodia Sanguine and Tinnea Aetiopica, which belong to Scutellarioideae and are related genera of Scutellaria.). The common genes of 27 chloroplast genomes (including S. franchetiana) were extracted and concatenated with Phylosuite (Zhang et al. Citation2020). These sequences including 68 common genes were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT, v7.450) (Rozewicki et al. Citation2019), and a phylogenetic tree was constructed using the alignment and maximum-likelihood (ML) method implemented in IQtree with the parameter ‘GTR + F+R2’ (Trifinopoulos et al. Citation2016).
Result
The total length of the complete chloroplast genome of S. franchetiana (GenBank accession number: OQ909432.1) is 151,852 bp with 38.37% GC content (). The chloroplast genome of S. franchetiana contained a typical quadratic structure including a pair of inverted repeats (IRs) of 25,219 bp separated by a single-copy (SSC) region of 17,520 bp and a single-copy (LSC) region of 83,894 bp. The chloroplast genome included 130 genes, including 86 protein-coding genes, eight ribosome RNA (rRNA), and 36 transfer RNA (tRNA) genes. Nineteen of the protein-coding genes have one intron and four have two introns. The ML tree showed that the phylogenetic relationship of S. franchetiana was closest to Scutellaria tsinyunensis and Scutellaria microviolacea (). The chloroplast genome sequence of S. franchetiana is a valuable resource for studying the phylogenetic evolution of the genus Scutellaria.
Figure 3. The complete chloroplast genomes of S. franchetiana and 24 other Scutellaria species were used to infer phylogenetic trees by maximum likelihood method. Holmskiodia Sanguine and Tinnea Aetiopica were selected as outgroups. With 1000 times as the bootstrap value, numbers above the branch represent the bootstrap value.
Discussion and conclusion
The complete chloroplast genome of S. franchetiana was first sequenced and found to exhibit a total length of 152,050 bp. The genome size and gene content of S. franchetiana is not significantly different from those of most chloroplast genomes or plastomes in the genus Scutellaria (Chen Citation2019; Shan et al. Citation2021). At present, there is little research on the phylogeny of Scutellaria, mainly because there are few chloroplasts (25 complete chloroplast genomes) and mitochondrial genomes (only S. tsinyunensis) published. The close relationship between S. franchetiana and S. tsinyunensis can provide reference for studying the phylogenetic relationship of the whole genus. Therefore, a more complete Scutellaria species chloroplast sequence is necessary for further research on phylogenetic relationships in Scutellaria genus. The exploration of molecular markers targeting the highly variable regions of plastid genome, such as ycf1, should be helpful to investigate systematic evolution of Scutellaria genus. The unveiling of the chloroplast genome sequence of S. franchetiana will provide meaningful information for analyzing the genetic diversity and phylogenetic relationships in Scutellaria species.
Authors’ contributions
Jiabin Yu and Ning Huang analyzed the sequence data and drafted the paper. Chuliang Wang designed this work and reviewed the work critically for important intellectual content. Jutao Sun collected the specimen material and conducted the experiment. Weikai Gao agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethical approval
The authors declare no ethical or legal violations when obtaining the study materials and performing the research. The species used in this study is not listed on the IUCN Red List, and the sample was legally collected by guidelines stipulated in national and international regulations. The materials were collected in a location not designated as a protected area in China.
Supplemental Material
Download MS Word (2.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The supported genome sequence data for the findings of this study can be obtained from GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) with the accession number of OQ909432.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA947610, SAMN33862474, SRR23952261, respectively.
References
- Bruno M, Piozzi F, Maggio AM, Simmonds MS. 2002. Antifeedant activity of neoclerodane diterpenoids from two Sicilian species of Scutellaria. Biochem Syst Ecol. 30(8):793–799. doi:10.1016/S0305-1978(01)00143-0.
- Chen S. 2019. Genetic and phylogenetic analysis of the complete genome for the herbal medicine plant of Scutellaria baicalensis from China. Mitochondrial DNA B Resour. 4(1):1683–1685. doi:10.1080/23802359.2019.1605859.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Islam MA, Zilani MNH, Biswas P, Khan DA, Rahman MH, Nahid R, Nahar N, Samad A, Ahammad F, Hasan MN, et al. 2022. Evaluation of in vitro and in silico anti-inflammatory potential of some selected medicinal plants of Bangladesh against cyclooxygenase-II enzyme. J Ethnopharmacol. 285:114900. doi:10.1016/j.jep.2021.114900.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM, McVean G, Lunter G; WGS500 Consortium. 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 46(8):912–918. doi:10.1038/ng.3036.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10. doi:10.1093/nar/gkz342.
- Shan Y, Pei X, Yong S, Li J, Qin Q, Zeng S, Yu J. 2021. Analysis of the complete chloroplast genomes of Scutellaria tsinyunensis and Scutellaria tuberifera (Lamiaceae). Mitochondrial DNA B Resour. 6(9):2672–2680. doi:10.1080/23802359.2021.1920491.
- Shen J, Li P, Liu S, Liu Q, Li Y, Sun Y, He C, Xiao P.,. 2021. Traditional uses, ten-years research progress on phytochemistry and pharmacology, and clinical studies of the genus Scutellaria. J Ethnopharmacol. 265:113198. doi:10.1016/j.jep.2020.113198.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Trifinopoulos J, Nguyen L T, von Haeseler A, et al. 2016. W-IQ-T REE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1): W232–W235.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
