Abstract
The lowland paca (Cuniculus paca) is a nocturnal, widespread, and solitary large-sized rodent in the family Cuniculidae, and one of the most frequently hunted mammals in the Neotropical forests of Latin America. We assembled the first complete mitochondrial genome of lowland paca using three closely related hystricognath species as reference sequences. The mitochondrial genome is 16,770 basepairs (bp) in length, with similar characteristics of vertebrate mitochondrial genomes. We performed phylogenetic analyses using 26 mitochondrial genome of hystricognath species based on thirteen protein-coding genes. The result confirms the taxonomical placement among the New World hystricognath rodents with high support. The placement is consistent with previous phylogenetic studies based on individual mitochondrial and nuclear genes. The current study improves the phylogenic resolution of hystricognath rodents.
1. Introduction
The lowland paca (Cuniculus paca, Linnaeus, 1766) is the living species of Cuniculus, the only genus in the family Cuniculidae. The pacas were previously placed among the Agouti species in the family Dasyproctidae, genus Dasyprocta, subfamily Agoutinae, but has been defined as an independent family due to morphological differences (Grzimek Citation2003). Lowland pacas () are nocturnal, herbivorous and solitary large-sized (6–12 kg) rodents with a relatively low fecundity for rodents (Mayor et al. Citation2013, Citation2017). The species inhabit a variety of forest types in tropical areas within Central and South America from eastern and southern Mexico to northeast Argentina (Patton et al. Citation2015). Introductions into Cuba and the Lesser Antilles have occurred recently (Patton et al. Citation2015). Although this species is currently the most hunted Neotropical rodent species (El Bizri, Morcatty, Ferreira, et al. Citation2020; El Bizri, Morcatty, Valsecchi, et al. Citation2020), wild populations are stable and are listed as Least Concern by The IUCN Red List of Threatened Species in 2016 (Emmons Citation2016).
2. Materials and methods
A volume of ∼100 µl blood sample was collected by local hunters in the Yavarí-Mirin River basin (04°19′53″ S, 71°57′33″ W) in the Peruvian Amazon and was spotted on Whatman FTA card for storage. The specimen and DNA isolate were deposited in the frozen sample archive of Leibniz Institute for Zoo and Wildlife Research (IZW, https://www.izw-berlin.de/en/home.html; contact: Alex D. Greenwood, [email protected]) under the voucher number 151310178. We sliced the FTA card, which was diluted using AVL buffer in QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). After 10 min of vortex at 2000 rpm, genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Illumina DNA sequencing library following Meyer and Kircher (Citation2010) was built with an average insert size of 300 bp, and was subsequently sequenced on the Illumina MiSeq platform using MiSeq Reagent Kit v2 (PE, 2 × 150 cycles).
We removed adapter sequences with Cutadapt v.1.15 (Martin Citation2011) and trimmed low quality reads with Trimmomatic v.0.38 (Bolger et al. Citation2014). Complete circular mitochondrial genome was assembled with MitoFinder (Allio et al. Citation2020) by using three complete mitogenomes of hystricognath species as reference sequences: Cavia aperea (NC_046949), Cavia porcellus (NC_000884), and Coendou insidiosus (NC_021387). All reads were then mapped backed to the de novo assembled mitogenome sequence using BWA v. 0.7.17 (Li and Durbin Citation2009). We called consensus sequence with default parameters using bcftools v.1.7 (Danecek and McCarthy Citation2017). The resultant mitochondrial genome was annotated using MitoFinder (Allio et al. Citation2020) after reverse complementing.
A phylogenetic analysis was performed using 13 PCGs of the lowland paca and 26 hystricognath rodents. Each gene from different species was aligned individually using MAFFT (-linsi) v7.310 (Katoh and Standley Citation2013). The multiple sequence alignments of the 13 genes were concatenated using catfasta2phyml (https://github.com/nylander/catfasta2phyml). A maximum-likelihood tree was estimated using IQ-TREE v.2.0.3 (Minh et al. Citation2020) with 1000 bootstrap replicates. We applied TVM + I + G as the substitution model for ND6 gene and GTR + I + G for other 12 genes. The substitution models were determined by PartitionFinder v.2.1.1 (Lanfear et al. Citation2016).
3. Results
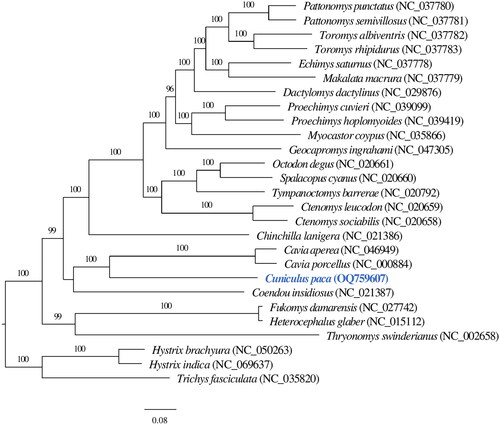
In total, 45,552 merged reads out of 1,658,046 read pairs were mapped back to the assembled mitogenome sequence. The mitochondrial genome sequence has 100% total coverage with an average of ×370 base coverage, ranging from 28× to 1890× (Supplementary Figure 1). The complete mitochondrial genome of the lowland paca is 16,770 bp length with GC content of 40.81%, constituting a control region (position: 15,585–16,770) and a set of 37 genes: 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes (). The nucleotides of the mitogenome are composed of 32.98% adenine (A), 26.21% thymine (T), 13.33% guanine (G), and 27.47% cytosine (C). The majority of the PCGs initiate with the common vertebrate start codon ATG, except ND2 and ND3 with ATT, and ND5 and ND6 with ATC. Ten PCGs terminate with stop codons either TAG (ND1, ND2, COX1, ATP8, and ND3) or TAA (COX2, ATP6, ND4L, ND5, and ND6), while CYTB gene terminates with arginine coding codon AGA, COX3 with a single T and ND4 with TA. Phylogenetic analysis shows the lowland paca (Cuniculus paca) mitogenome clusters with the genus Cavia and is placed in the infraorder Hystricognathi within the hystricognath rodents ().
Figure 2. Clockwise view of the mitochondrial genome of the Cuniculus paca generated from CGView (Stothard and Wishart Citation2005).
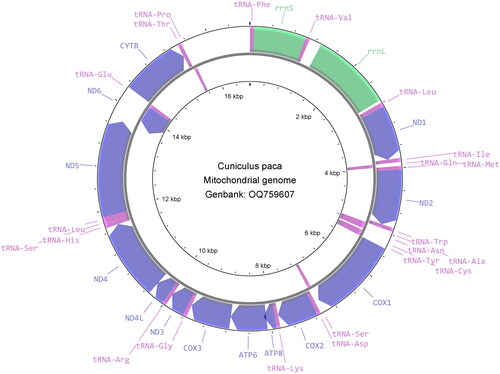
Figure 3. The phylogenetic relationship of lowland paca (Cuniculus paca) with 26 hystricognath rodent species are inferred from maximum-likelihood estimation based on 13 protein-coding genes with 1000 bootstrap replicates. Support values were given above each branch. The NCBI accession of mitogenome references used was displayed following species scientific names in parenthesis. The tip marked in blue depicts the mitochondrial genome of Cuniculus paca generated in this study.
4. Discussion and conclusions
Phylogenetic analysis confirms the taxonomical position of lowland paca (Cuniculus paca) mitogenome in the infraorder Hystricognathi within the hystricognath rodents, showing that Cuniculus paca is closely related to the genus Cavia (). The results are consistent with previous analyses based on multiple mitochondrial and nuclear genes (Voloch et al. Citation2013). The current study de novo assembles a mitochondrial genome of lowland paca (Cuniculus paca) and improves the phylogenic resolution of hystricognath rodents.
Author contributions
ADG, PM, MLSR and JL developed the project. JL developed and conducted the laboratory and bioinformatics workflow. PM and MLSR helped the project management and sample collection in Peru, and provided the expertise about the species. JL, ADG, and PM wrote the article.
Ethics statement
Research protocols for NHPs sampling were approved by the Peruvian Forestry and Wildlife Agency (No. 258-2019-MINAGRI-SERFOR-DGGSPFFS) and the Institutional Animal Use Ethics Committee of the Universidad Peruana Cayetano Heredia (ref. SIDISI 102142).
Supplemental Material
Download PNG Image (336.5 KB)Acknowledgements
This work has received financial support from the ERANet-LAC [Grant No. 17/HLH-0271] within the research project [Contract No. 136-2018-FONDECYT]. The authors thank Dorina Meneghini for providing bioinformatic support. JL acknowledges the support of the China Scholarship Council. Finally, the authors would like to acknowledge local people of Nueva Esperanza in the Yavari-Mirin River for their help in collecting biological samples.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The mitochondrial genome sequence supporting this study is publicly released at NCBI GenBank with the accession no. OQ759607. The associated sequencing data are deposited at NCBI Sequencing Reads Archive (SRA) under the BioProject PRJNA950048 (accession number SRR24006478 and BioSample number SAMN33969573).
Additional information
Funding
References
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905. doi: 10.1111/1755-0998.13160.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. doi: 10.1093/bioinformatics/btu170.
- Danecek P, McCarthy SA. 2017. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics. 33:2037–2039. doi: 10.1093/bioinformatics/btx100.
- El Bizri HR, Morcatty TQ, Ferreira JC, Mayor P, Vasconcelos Neto CFA, Valsecchi J, Nijman V, Fa JE. 2020. Social and biological correlates of wild meat consumption and trade by rural communities in the Jutaí River Basin, Central Amazonia. J Ethnobiol. 40(2):183–201. doi: 10.2993/0278-0771-40.2.183.
- El Bizri HR, Morcatty TQ, Valsecchi J, Mayor P, Ribeiro JES, Vasconcelos Neto CFA, Oliveira JS, Furtado KM, Ferreira UC, Miranda CFS, et al. 2020. Urban wild meat consumption and trade in central Amazonia. Conserv Biol. 34:438–448. doi: 10.1111/cobi.13420.
- Emmons L. 2016. Cuniculus paca. The IUCN Red List of Threatened Species 2016: e.T699A22197347.
- Grzimek B. 2003. Grzimek’s animal life encyclopedia, volume 16: mammals V. Farmington Hills (MI): Gale Group.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. doi: 10.1093/molbev/mst010.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773. doi: 10.1093/molbev/msw260.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760. doi: 10.1093/bioinformatics/btp324.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. doi: 10.14806/ej.17.1.200.
- Mayor P, El Bizri H, Bodmer RE, Bowler M. 2017. Assessment of mammal reproduction for hunting sustainability through community-based sampling of species in the wild. Conserv Biol. 31(4):912–923. doi: 10.1111/cobi.12870.
- Mayor P, Guimaraes DA, Lopez C. 2013. Functional morphology of the genital organs in the wild paca (Cuniculus paca) female. Anim Reprod Sci. 140:206–215. doi: 10.1016/j.anireprosci.2013.06.010.
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010:pdb.prot5448. doi: 10.1101/pdb.prot5448.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. doi: 10.1093/molbev/msaa015.
- Patton JLP, Pardiñas UFJ, D’Elía G. 2015. Mammals of South America, volume 2: rodents. Chicago: University of Chicago Press.
- Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics. 21(4):537–539. doi:10.1093/bioinformatics/bti054.
- Voloch CM, Vilela JF, Loss-Oliveira L, Schrago CG. 2013. Phylogeny and chronology of the major lineages of New World hystricognath rodents: insights on the biogeography of the Eocene/Oligocene arrival of mammals in South America. BMC Res Notes. 6(1):160. doi: 10.1186/1756-0500-6-160.