Abstract
Pseudofabraea citricarpa (Dermateaceae: Helotiales) is known as a significant pathogen causing Citrus target spot disease and results in profound yield loss. In the present study, the complete mitochondrial genome (mitogenome) determined based on next-generation sequencing technology. The circular mitogenome (56,935 bp) comprised 14 conserved protein-coding genes (PCGs), 16 ORFs, two ribosomal RNA genes (rns and rnl), one non-coding RNA gene (rnpB), one ribosomal protein S3 (rps3) and 28 transfer RNA (tRNA) genes. The overall base composition is as follows: 36.08% A, 35.25% T, 13.04% C, and 15.63% G, with a GC content of 28.70%. The phylogenetic analysis shows that P. citricarpa, belonging to Dermateaceae, forms a separate clade and is sister to Sclerotiniaceae. The mitogenome of P. citricarpa reported in this study provides more molecular data for further research on the evolutionary relationships of Helotiales.
Introduction
Pseudofabraea citricarpa (Zhu et al.) Chen et al. (Citation2016) infect Citrus spp. causing target spot, was described as a fungal disease in 2012 (Zhu et al. Citation2012). The disease was first reported on Citrus unshiu Marcow (1921) in Chenggu county, Shaanxi province, China (Xiao et al. Citation2021). A large number of citrus trees fell ill or died, the yield declined sharply, and some orchards were destroyed, which severely restricted the local economic development due to citrus target spot (Yang et al. Citation2019). In our present survey, the species distributed in Shaanxi, Chongqing, Hunan, and Hubei provinces of China, tending to invade the upper reaches of the Yangtze River. The citrus target spot pathogen initially identified as Cryptosporiopsis citricarpa Zhu et al. (Citation2012) based on Koch’s postulates and molecular phylogenetic and morphological characteristics (Zhu et al. Citation2012). Then, the species was recombined into P. citricarpa (Chen et al. Citation2016). The fast detection method for the citrus target spot pathogen was developed based on a SCAR mark (Yang et al. Citation2018). The critical pathogenicity factors of P. citricarpa involved in the induction of citrus target spots were determined by comparative transcriptomic and secretomic analyses (Yang et al. Citation2019). However, little known about the genetic diversity and genomic information of P. citricarpa. This study aims to describe the mitochondrial genomes (mitogenome) of P. citricarpa, to provide insight into the genetics and evolutionary biology of P. citricarpa.
Materials and methods
Strains of P. citricarpa were isolated from citrus target spot leaves in the Yiling District, Yichang, Hubei province, China (111°0′27.308″E, 30°46′42.642″N) (). The strain YLSDP74 was deposited at Food Crops Research Institute, Yunnan Academy of Agricultural Sciences, China (https://www.yaas.org.cn, Mingliang Ding, [email protected]).
Figure 1. Symptoms of Citrus target spot on C. unshiu caused by Pseudofabraea citricarpa. Photographed by Quan Chen on 18 January 2021.

Mycelia cultured at 25 °C for 15 days before sequencing. The total genomic DNA of P. citricarpa was extracted using the Ezup Column Fungi Genomic DNA Purification Kit. Whole-genome sequencing was performed at the Cloud Health Medical Group Ltd using Illumina XTen sequencing system (Illumina Inc., San Diego, CA). The mitogenome of P. citricarpa assembled and annotated using the software GetOrganelle version 1.7.5 and MFannot tools, respectively (Utomo et al. Citation2019; Jin et al. Citation2020). Predictive annotations of tRNA genes was conducted using tRNAscan-SE 2.0.10 software (Chan and Lowe Citation2019). The OGDRAW version 1.3.1 (Greiner et al. Citation2019) used to draw the mitogenomic circular map.
To investigate the phylogenetic relationship of P. citricarpa to other 14 fungal species in Helotiales, and 16 mitogenomes were downloaded from the NCBI database and combined with the P. citricarpa for phylogenetic analysis. Pseudocercospora fijiensis (M. Morelet) Deighton (1976) and Pseudocercospora mori (Hara) Deighton (1976) designated as the outgroup. The phylogenetic trees constructed using RAxML v 8.2.12 (Alexandros Citation2014) with the optimal model GTR + F + I + I + R3 and 1000 rapid bootstrap replication on 13 PCGs.
Results
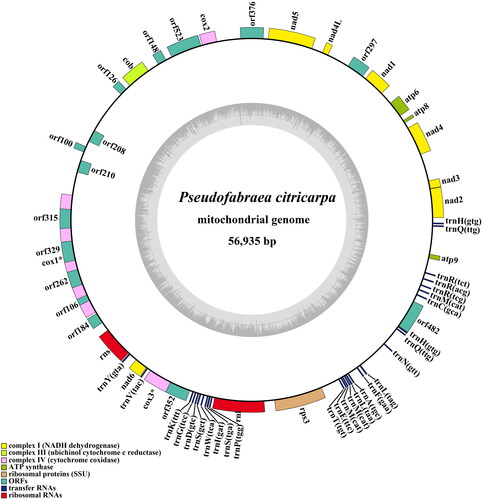
The complete mitogenome sequence of P. citricarpa deposited in GenBank (accession no. ON960225). The average coverage depth registered an impressive ×901, accompanied by a minimum depth of ×327 and a substantial maximum depth of ×1891 (Supplementary Figure S1). The circular genome (56,935 bp) comprised of 14 conserved protein-coding genes (PCGs), 16 ORFs (orf100, orf106, orf126, orf148, orf184, orf208, orf210, orf262, orf297, orf315, orf329, orf352, orf376, orf482, orf523, and orf812), two ribosomal RNA genes (rns and rnl), one non-coding RNA gene (rnpB), one ribosomal protein S3 (rps3), and 28 transfer RNA (tRNA) genes (). The 14 PCGs respectively encoded the seven ubiquinone reductase subunits of NADH (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), three cytochrome oxidase subunits (cox1, cox2, and cox3), three ATP synthase subunits (atp6, atp8, and atp9), and the apocytochrome b (cob). The total length of 14 PCGs and 16 ORFs was 27,744 bp, which accounted for 48.73% of the whole mitogenome. The 28 tRNA genes (trnH(gtg), trnQ(ttg), trnR(tct), trnR(acg), trnR(tcg), trnM(cat), trnC(gca), trnH(gtg), trnQ(ttg), trnN(gtt), trnL(tag), trnF(gaa), trnA(tgc), trnL(taa), trnM(cat), trnM(cat), trnE(ttc), trnT(tgt), trnP(tgg), trnS(tga), trnI(gat), trnW(tca), trnS(gct), trnD(gtc), trnG(tcc), trnK(ttt), trnV(tac), and trnY(gta)) ranged in size from 70 bp to 86 bp. The overall base composition was as follows: 36.08% A, 35.25% T, 13.04% C, and 15.63% G, with a GC content of 28.70%. Phylogenetic analysis showed that P. citricarpa within Dermateaceae was close to species of Sclerotiniaceae ().
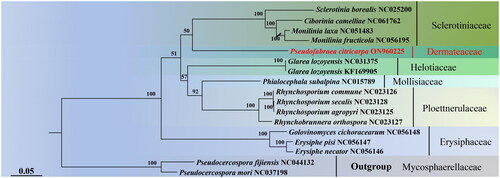
Figure 3. Maximum-likelihood tree generated using 13 concatenated mitochondrial protein-coding genes (atp6, atp8, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) from Pseudofabraea citricarpa and 16 other fungal species. The following sequences were used: Sclerotinia borealis (NC_025200) (Mardanov et al. Citation2014), Ciborinia camelliae (NC_061762) (Valenti et al. Citation2021), Monilinia laxa (NC_051483) (Yildiz and Ozkilinc Citation2020), Monilinia fructicola (NC_056195), Glarea lozoyensis (NC_031375) (Zhang et al. Citation2017), Glarea lozoyensis (KF_169905) (Youssar et al. Citation2013), Phialocephala subalpine (NC_015789) (Duò et al. Citation2012), Rhynchosporium commune (NC_023126), Rhynchosporium secalis (NC_023128), Rhynchosporium agropyri (NC_023125), Rhynchosporium orthosporum (NC_023127) (Torriani et al. Citation2014), Golovinomyces cichoracearum (NC_056148), Erysiphe pisi (NC_056147), Erysiphe necator (NC_056146) (Zaccaron and Stergiopoulos Citation2021), Pseudocercospora fijiensis (NC_044132) (Arcila-Galvis et al. Citation2021), and Pseudocercospora mori (NC_037198).
Discussion and conclusions
The mitogenome of P. citricarpa consists of 14 PCGs, 16 ORFs, rns, rnl, rnpB, rps3, and 28 tRNA. Among the 14 PCGs, the longest gene is nad5 (2247 bp), while the shortest is atp8 (147 bp). The mitogenome of P. citricarpa exhibits a significantly higher proportion of AT nucleotides compared to those of Sclerotinia, Monilinia, and Ciborinia (Mardanov et al. Citation2014; Medina et al. Citation2020; Yildiz and Ozkilinc Citation2020; Valenti et al. Citation2021). The phylogenetic tree confirms the position of Dermateaceae within Helotiales and provides robust molecular evidence for the classification of Pseudofabraea species. Several provinces in China have reported citrus target spot, causing significant harm to the citrus industry (Chen et al. Citation2023). In confronting this challenge effectively, leveraging the capabilities of molecular markers derived from the P. citricarpa mitochondrial genome holds the promise of refining the accuracy in pinpointing and monitoring the extent and severity of citrus target spot disease. This endeavor promises to yield crucial revelations for timely disease detection and proactive preventive approaches, thereby establishing a resilient foundation for orchestrating disease surveillance and control endeavors.
Author contributions
Conceived and designed the study: Hui Li. Performed the experiments: Songlin Xu and Mei Yang. Analyzed the data: Wenjing Zhang and Zhengang Duan. Draft the manuscript: Jinhui He. Revised the manuscript: Jinhui He. Final approval of the version to be published: Mingliang Ding and Quan Chen. All authors agreed to be accountable for all aspects of the work.
Ethics statement
The study did not involve humans or animals. In this study, samples of citrus target spot disease can be collected without ethical approval or permission.
Supplemental Material
Download MS Word (152.8 KB)Supplemental Material
Download EPS Image (1.6 MB)Supplemental Material
Download JPEG Image (80.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ON960225.1/ under the reference number ON960225. The associated “BioProject”, “Bio-Sample”, and “SRA” numbers are PRJNA917114, SAMN32522014, and SRR22953112, respectively.
Additional information
Funding
References
- Alexandros S. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Arcila-Galvis JE, Arango RE, Torres-Bonilla JM, Arias T. 2021. The mitochondrial genome of a plant fungal pathogen Pseudocercospora fijiensis (Mycosphaerellaceae), comparative analysis and diversification times of the Sigatoka disease complex using fossil calibrated phylogenies. Life. 11(3):215. doi: 10.3390/life11030215.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1.
- Chen C, Verkley GJ, Sun GY, Groenewald JZ, Crous PW. 2016. Redefining common endophytes and plant pathogens in Neofabraea, Pezicula, and related genera. Fungal Biol. 120(11):1291–1322. doi: 10.1016/j.funbio.2015.09.013.
- Chen Q, Zhang WJ, Xu SL, Li H, He JH. 2023. Occurrence dynamics of citrus target spot in Wanzhou district of Chongqing city and compound fungicide screening of Pseudofabraea citricarpa. BIO Web Conf. 60:01021. doi: 10.1051/bioconf/20236001021.
- Duò A, Bruggmann R, Zoller S, Bernt M, Grünig CR. 2012. Mitochondrial genome evolution in species belonging to the Phialocephala fortinii s.l. – Acephala applanata species complex. BMC Genomics. 13(1):166. doi: 10.1186/1471-2164-13-166.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Mardanov AV, Beletsky AV, Kadnikov VV, Ignatov AN, Ravin NV. 2014. The 203 kbp mitochondrial genome of the phytopathogenic fungus Sclerotinia borealis reveals multiple invasions of introns and genomic duplications. PLOS One. 9(9):e107536. doi: 10.1371/journal.pone.0107536.
- Medina R, Franco MEE, Bartel LC, Martinez AV, Saparrat MCN, Balatti PA. 2020. Fungal mitogenomes: relevant features to planning plant disease management. Front Microbiol. 11:978. doi: 10.3389/fmicb.2020.00978.
- Torriani SFF, Penselin D, Knogge W, Felder M, Taudien S, Platzer M, McDonald BA, Brunner PC. 2014. Comparative analysis of mitochondrial genomes from closely related Rhynchosporium species reveals extensive intron invasion. Fungal Genet Biol. 62:34–42. doi: 10.1016/j.fgb.2013.11.001.
- Utomo C, Tanjung ZA, Aditama R, Buana RFN, Pratomo ADM, Tryono R, Liwang T. 2019. Complete mitochondrial genome sequence of the phytopathogenic basidiomycete Ganoderma boninense strain G3. Microbiol Resour Announc. 8(5):e00968-18. doi: 10.1128/MRA.00968-18.
- Valenti I, Degradi L, Kunova A, Cortesi P, Pasquali M, Saracchi M. 2021. The first mitochondrial genome of Ciborinia camelliae and its position in the Sclerotiniaceae family. Front Fungal Biol. 2:802511. doi: 10.3389/ffunb.2021.802511.
- Xiao XE, Zeng YT, Wen W, Cheng L, Qiao XH, Hou X, Li HY. 2021. First report and new hosts of Pseudofabraea citricarpa causing citrus target spot in china. Plant Health Progress. 22(1):26–30. doi: 10.1094/PHP-07-20-0056-RS.
- Yang YH, Fang AF, Yu Y, Bi CW, Zhou CY. 2019. Integrated transcriptomic and secretomic approaches reveal critical pathogenicity factors in Pseudofabraea citricarpa inciting citrus target spot. Microb Biotechnol. 12(6):1260–1273. doi: 10.1111/1751-7915.13440.
- Yang YH, Hu JH, Chen FJ, Ding DK, Zhou CY. 2018. Development of a SCAR marker-based diagnostic method for the detection of the citrus target spot pathogen Pseudofabraea citricarpa. Biomed Res Int. 2018:7128903. doi: 10.1155/2018/7128903.
- Yildiz G, Ozkilinc H. 2020. First characterization of the complete mitochondrial genome of fungal plant-pathogen Monilinia laxa which represents the mobile intron-rich structure. Sci Rep. 10(1):13644. doi: 10.1038/s41598-020-70611-z.
- Youssar L, Grüning BA, Günther S, Hüttel W. 2013. Characterization and phylogenetic analysis of the mitochondrial genome of Glarea lozoyensis indicates high diversity within the order Helotiales. PLOS One. 8(9):e74792. doi: 10.1371/journal.pone.0074792.
- Zaccaron AZ, Stergiopoulos I. 2021. Characterization of the mitochondrial genomes of three powdery mildew pathogens reveals remarkable variation in size and nucleotide composition. Microb Genom. 7(12):000720.
- Zhang YJ, Zhao YX, Zhang S, Chen L, Liu XZ. 2017. Reanalysis of the mitochondrial genome of the pneumocandin-producing fungus Glarea lozoyensis. Acta Microbiol Sin. 57(5):724–736.
- Zhu L, Wang XH, Huang F, Zhang JZ, Li HY, Ding DK, Hyde KD. 2012. A destructive new disease of citrus in China caused by Cryptosporiopsis citricarpa sp. nov. Plant Dis. 96(6):804–812. doi: 10.1094/PDIS-09-11-0775.