Abstract
Fargesia angustissima T. P. Yi, categorized into Arundinarieae (Poaceae: Bambusoideae), is a critical species endemic to Minshan Mountain, China. F. angustissima provides shelter and food sources for the giant panda and other endangered animals (e.g. red panda and snub-nosed monkey). This study assembled the complete chloroplast (cp) genome of F. angustissima using the high-throughput sequencing technique. The total cp length was 139,706 bp, containing 130 annotated genes with predicted GC content at 38.87%. The cp genome comprises two single-copy (LSC and SSC) regions, harboring 83,282 bp and 12,830 bp, respectively. The SSC regions were located between two inverted repeats (IR) regions (21,797 bp). Reconstruction of the phylogenetic tree illustrated that F. angustissima clustered F. canaliculata in Fargesia II. The study provides theoretical clues to explore the geographical distribution and species-level identification of the Fargesia genus.
Introduction
The F. angustissima T. P. Yi 1985 (You bamboo) was categorized into Arundinarieae, the subfamily of Bambusoideae, and distributed broadly to the Minshan Mountain, western Sichuan province, China. It shows a habitation on steep limestone slopes ranging from 800 to 1550 meters. F. angustissima is an evergreen, clump-forming woody bamboo with short rhizomes and erect-tall stems approximately 4 - 7 m (). The stiffer culm can be commonly utilized as pen resources and crutches for crop growth, while its split fibers are processed for weaving furniture and farming tools. Besides, as an indispensable component of the subtropical ecosystem, F. angustissima provides habitat and food for endangered animals, particularly giant pandas (Li et al. Citation2017). Given the significant morphological diversity (leaf sheaths and inflorescences) and complex evolutionary history (cold climatic conditions in the Quaternary) (Zhou et al. Citation2019), there remains uncertainty surrounding its long-standing taxonomic boundaries within the Fargesia genus (Guo and Li Citation2004; Ye et al. Citation2021). Whole plastome sequencing helps resolve complex evolutionary relationships among closely related species; however, limited information regarding the complete chloroplast (cp) genome is available in F. angustissima (Williams et al. Citation2016). In this study, the complete plastome of F. angustissima was deciphered using the Illumina high-throughput sequencing technique. The evolutionary tree reconstruction in Fargesia provides essential cues on population identification, conservation, and ecological application.
Materials
The fresh leaves of the F. angustissima were collected at Zhudu Park, Changning County (20°28'53ʺN, 105°52'21ʺE), Yibin City, Sichuan Province, China. Moreover, the voucher specimens (ID: ST20231110005) were deposited in the herbarium of Nanjing Forestry University (https://www.cfh.ac.cn/subsite/Albums.aspx?siteid=njfu), Nanjing, China (Tao Su, [email protected]).
Methods
The genomic DNA of F. angustissima was extracted by plant genomic DNA kit (Omega, Georgia, USA) and sequenced by the Novaseq platform (Illumina, San Diego, CA), following the manufacturer’s instructions. We perform library preparation and sequencing using next-generation sequencing (NGS) technology to extract and sequence total DNA from fresh leaves. The complete cp genome of F. qinlingensis (NC_040002) was used as a reference, followed by the genome assembly using the GetOrganelle (Jin et al. Citation2020). The assembled cp genome is subjected to annotation to determine the start positions and IR regions using Geseq (Tillich et al. Citation2017) and CpGAVAS (Liu et al. Citation2012). The genome annotation is performed on the identified genes. Furthermore, we corrected annotations by comparing them to ensure accuracy regarding F. qinlingensis. The cp genome map was drawn by the CPGview (http://www.1kmpg.cn/cpgview) (Liu et al. Citation2023). Finally, the complete cp sequence was submitted to GenBank (https://www.ncbi.nlm.nih.gov/) with the assigned accession ID OP821764.
An unrooted phylogenetic tree was constructed using the available complete cp sequence in GenBank to explore the evolutionary relationships between F. angustissima and the other 26 species in Poaceae. Multiple cp sequences were aligned using MAFFT (Katoh and Standley Citation2013). Then, MEGA X was applied to process the phylogenetic calculation by the Maximum Likelihood (ML) method with the Jones-Taylor-Thornton model based on 1000 rapid bootstraps (Kumar et al. Citation2018).
Results
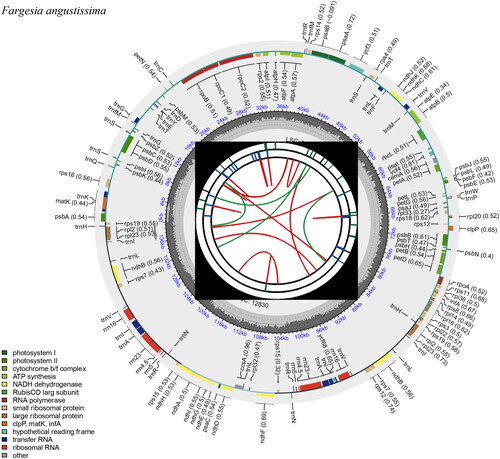
We have obtained a fully sequenced cp genome of F. angustissima that is 139,706 bp in sequence length and displays a typical circled quadripartite structure containing an 83,282 bp large single-copy (LSC) region, a 12,830 bp small single-copy (SSC) region (). While two inverted repeat regions (IRA and IRB) showed the same length of 21,797 bp with splicing genes deciphered (Figure S1 and S2). The GC content in the cp genome was 38.87%. The assembled cp sequence has similar features to other bamboo species within the Fargesia genus (Zhou et al. Citation2019). The F. angustissima cp genome was annotated with 130 unigenes, including 83 protein-encoding genes, thirty-nine transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes.
Figure 2. Gene map of the F. angustissima cp genome predicted by CPGView. From the center outward, the first track indicates the dispersed repeats; the second track shows the long tandem repeats as short blue bars; the third track shows the short tandem repeats or microsatellite sequences as short bars with different colors; the fourth track shows SSC, IRA and IRB, and LSC regions. The GC content along the genome is plotted on the fifth track, and annotated genes are shown on the sixth.
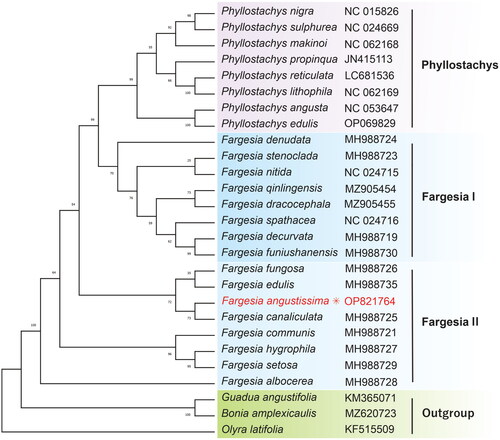
Based on the phylogenetic analyses, the unrooted evolutionary tree is divided into three subclades: Phyllostachys, Fargesia I, Fargesia II, and the Outgroup. The reconstruction of phylogenies revealed that F. angustissima clustered F. canaliculata within the Fargesia II group ().
Figure 3. Phylogenetic relationships of F. angustissima and other bamboo species. The unrooted evolutionary tree was constructed using the ML method involving 27 species in Bambusoideae. The sample and respected bootstrap support value were clustered for each branch node. The complete cp sequences and accession ID were used as follows: Phyllostachys nigra NC015826 (Zhang et al. Citation2011), P. sulphurea NC024669, P. propinqua JN415113 (Wu and Ge Citation2012), F. spathacea NC024716 and F. nitida NC024715 (Ma et al. Citation2014), F. denudata MH988724, F. decurvata MH988719, F. funiushanensis MH988730, F. stenoclada MH988723, F. fungosa MH988726, F. canaliculata MH988725, F. edulis MH988735, F. communis MH988721, F. albocerea MH988728, F. hygrophila MH988727, and F. setosa MH988729 (Zhou et al. Citation2019), Guadua anagustifolia KM365071 (Wu et al. Citation2015), and Olyra latifolia KF515509 (Burke et al. Citation2014).
Discussion and conclusion
In this study, we reported the complete cp genome of F. angustissima, displaying a sequence length of 139,706 bp, similar to other Fargesia genera (). The phylogenetic tree suggested consistency with the previous findings (Zhou et al. Citation2019), and the three clades (Phyllostachys, Fargesia I, and Fargesia II) also inferred the geographical distribution and altitude patterns of the classified Fargesia species in various growth areas. These results are compatible with the previous classification of the genus Fargesia, which solely relied on morphological characteristics, prompting difficulties in the classification at the species level (Zhou et al. Citation2019). Through cp genome inference, the molecular evolution of Fargesia appears to align with hypotheses about geographical and climatic factors. Hitherto, there are a total of 82 classified species in the NCBI database for the genus Fargesia. Nevertheless, only 18 complete cp genome sequencing data are publicly available in the Fargesia genus. Due to the lack of the cp genome of the Fargesia species in an accessible database, additional research is required to investigate the phylogeny of this genus. The current study provides theoretical clues to explore the geographical distribution and species-level identification of the Fargesia genus.
Author contributions
Tao Su and Hao Wu designed the research; Hao Wu, Xue Li, and Yingjian Zhang performed the research and analyzed the data; Xue Li, Hao Wu, and Xianglei Xu participated in the fieldwork; Hao Wu, Xue Li, and Yingjian Zhang wrote the draft. Tao Su revised and confirmed the manuscript. All authors agreed to be accountable for all aspects of the work.
Ethical approval
The materials used in this study were not related to protected species and were within the limits of the relevant national laws. We obtained permission from ZhuDu Park to collect plant specimens.
Supplemental Material
Download MS Word (34.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting this study’s findings are available in GenBank of NCBI under accession No. OP821764.1. The associated Bio-Project, Bio-Sample, and SRA numbers are PRJNA900166, SAMN31680877, and SRR22253535, respectively.
Additional information
Funding
References
- Burke SV, Clark LG, Triplett JK, Grennan CP, Duvall MR. 2014. Biogeography and phylogenomics of New World Bambusoideae (Poaceae), revisited. Am J Bot. 101(5):886–891. doi: 10.3732/ajb.1400063.
- Guo Z-H, Li D-Z. 2004. Phylogenetics of the Thamnocalamus group and its allies (Gramineae: Bambusoideae): inference from the sequences of GBSSI gene and ITS spacer. Mol Phylogenet Evol. 30(1):1–12. doi: 10.1016/s1055-7903(03)00161-1.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Li G, Song H, Altigani LAA, Zheng X, Bu S. 2017. Changes of foraging patch selection and utilization by a giant panda after bamboo flowering. Environ Sci Pollut Res Int. 24(19):16418–16428. doi: 10.1007/s11356-017-9164-5.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715. doi: 10.1186/1471-2164-13-715.
- Ma P-F, Zhang Y-X, Zeng C-X, Guo Z-H, Li D-Z. 2014. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (poaceae). Syst Biol. 63(6):933–950. doi: 10.1093/sysbio/syu054.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. 2016. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol Phylogenet Evol. 96:1–8. doi: 10.1016/j.ympev.2015.11.021.
- Wu M, et al. 2015. The complete chloroplast genome of Guadua angustifolia and comparative analyses of neotropical-paleotropical bamboos. PloS One. 10:e0143792. doi: 10.1371/journal.pone.0143792.
- Wu Z-Q, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62(1):573–578. doi: 10.1016/j.ympev.2011.10.019.
- Ye X-Y, Zhang Y-X, Li D-Z. 2021. Two new species of Yushania (Poaceae: bambusoideae) from South China, with a taxonomic revision of related species. Plant Divers. 43(6):492–501. doi: 10.1016/j.pld.2021.03.001.
- Zhang Y-J, Ma P-F, Li D-Z. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PloS One. 6(5):e20596. doi: 10.1371/journal.pone.0020596.
- Zhou Y, Zhang Y-Q, Xing X-C, Zhang J-Q, Ren Y. 2019. Straight from the plastome: molecular phylogeny and morphological evolution of Fargesia (Bambusoideae: Poaceae). Front Plant Sci. 10:981. doi: 10.3389/fpls.2019.00981.

