Abstract
Ocimum basilicum L. var. basilicum (Sweet Basil) is an aromatic herb belonging to the family Lamiaceae and is known for its medicinal uses. It is commonly used in traditional medicine for its therapeutic value, including anti-allergic, anti-inflammatory, antioxidant, antitumor, and antimicrobial properties. In this study, we generated the complete chloroplast genome sequence of O. basilicum var. basilicum using Illumina paired-end sequencing data. The chloroplast genome was 152,407 bp in length, containing a large single-copy (LSC) region of 83,409 bp and a small single-copy region (SSC) of 17,604 bp, separated by a pair of inverted repeats (IRs) of 25,697 bp. The genome contained 134 genes, including 89 protein-coding, 37 tRNA, and eight rRNA genes. Nine genes had one intron, two genes had two introns, and others did not have any intron. Overall GC content of the chloroplast genome was 38%, while that of LSC, SSC, and IR regions was 35.9%, 31.6%, and 43.1%, respectively. Phylogenetic analysis of the chloroplast genomes revealed that O. basilicum var. basilicum was closely related to Ocimum basilicum from the Ocimum species.
Introduction
Ocimum basilicum L. is used in traditional medicine for treating lung and respiratory disorders (Aminian et al. Citation2021). The leaves and flowers of sweet basil are used to treat insect stings, snake bites, and skin illnesses (Rudayni et al. Citation2021). The roots and seeds were used to treat bleeding piles, respiratory disorders, hemorrhage, and fever. The estimated consumption of the dried O. basilicum plants in herbal medicine was 1000–2000 MT. It is possibly used as a substitute or adulterant in Ocimum americanum because of their common trade names, ‘Kali tulsi’ and ‘Ban tulsi’ (Ravikumar et al. Citation2018). O. basilicum is abundant in phytochemicals compounds such as alkaloids, tannins, flavonoids, and saponins, which possess anti-inflammatory, antioxidant, antiviral, and antimicrobial activities (Shahrajabian et al. Citation2020; Kurnia et al. Citation2023). Six varieties were described in O. basilicum, and the morphological variations among the varieties of O. basilicum were well-studied (Rawat et al. Citation2016). In this study, we describe the chloroplast genome of one of the varieties, O. basilicum var. basilicum. It is a shrub that grows up to 97.5 cm in height. Leaves are elliptic to egg-shaped and are up to 8 cm long. The stem and lamina are purple to green in color (Rawat et al. Citation2016). The pubescence on the stem and lamina is medium. The color of the flowers is pinkish-white. In recent years, the complete chloroplast genomes of Ocimum species have been reported (Balaji et al. Citation2021; Harini et al. Citation2021; Kavya et al. Citation2021). However, the complete chloroplast genome of Ocimum basilicum variety basilicum remains to be studied. In this study, we sequenced the chloroplast genome of O. basilicum var. basilicum, which provides information for further taxonomic and DNA barcoding studies among the Ocimum species.
Materials and methods
Plant material of O. basilicum var. basilicum () was collected from Pallavaram, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°58′01.4″N 80°08′42.3″E). The herbarium specimen was prepared and authenticated by the taxonomist (Dr. Senthilkumar Umapathy, Department of Botany, Madras Christian College, Tambaram, Tamil Nadu, India; email: [email protected]). The voucher specimen was deposited under the accession number MH178193 in the Madras Herbarium (MH) of the Botanical Survey of India, Southern Regional Centre, T.N.A.U. Campus, Coimbatore, Tamil Nadu, India (https://bsi.gov.in/regional-centres/en?rcu=133, Dr. M. U. Sharief, Scientist-F and Head of Office, email: [email protected]). Total genomic DNA from O. basilicum var. basilicum was extracted using the CTAB method (Doyle and Doyle Citation1987), with minor modifications (Balaji and Parani Citation2022). From the genomic DNA, a paired-end DNA library was constructed using the Nextera XT Library Prep Kit (Cat. No. FC-131-1024), according to the manufacturer’s protocol. The library was sequenced on the Illumina Novoseq 6000 platform (Illumina Inc., San Diego, CA) with a paired-end sequencing length of 150 bp. We generated 3.15 Gb of data with more than 86% q30 bases. The complete chloroplast genome of O. basilicum var. basilicum was assembled using the program GetOrganelle v1.7.5 (Jin et al. Citation2020), with O. basilicum chloroplast genome (NC_035143.1) as a reference. The assembled chloroplast genome of Ocimum basilicum var. basilicum was annotated with GeSeq (Tillich et al. Citation2017). The predicted transfer RNAs (tRNAs) were identified by tRNAscan-SE 2.0 (Lowe and Chan Citation2016). In addition, the CPGVIEW (www.1kmpg.cn/cpgview/) (Liu et al. Citation2023) was applied to structures to visualize the intron-containing genes. The sequencing depth of assembled chloroplast genome was done by aligning to the raw reads using a BWA aligner (Li and Durbin Citation2009). The bam file was viewed using the software Qualimap to obtain the coverage map (Fernando et al. Citation2012). The phylogenetic analysis was performed based on the complete chloroplast genome sequences of 20 species from the Lamiales order. A maximum-likelihood tree was generated using 1000 bootstrap replicates, and the best substitution model of Timura-3 parameter, G + I, in MEGA version 11.0.13 (Tamura et al. Citation2021) from the alignments created using the MAFFT program (Katoh and Standley Citation2013).
Results and discussion
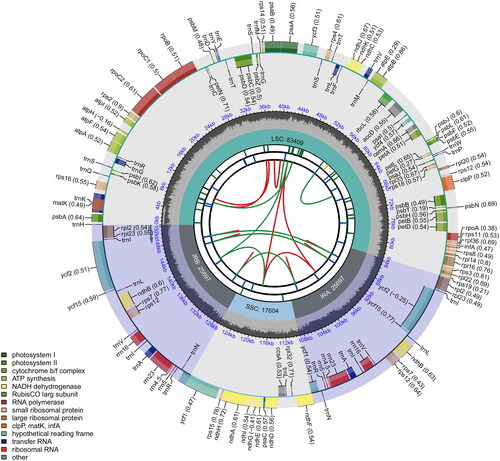
The chloroplast genome sequence of O. basilicum var. basilicum was 152,407 bp with a mean coverage of 446× (). It showed a typical quadripartite structure, including a large single-copy (LSC) region of 83,409 bp, a small single-copy (SSC) region of 17,604 bp, and a pair of inverted repeats (IRs) of 25,697 bp. The chloroplast genome contained 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16) were single-intron genes, and two genes (ycf3, clpP1) had two introns (Supplementary Figures 1 and 2). The overall GC content was 38%, while LSC, SSC, and IR regions were 35.9%, 31.6%, and 43.1%, respectively. The average depth of assembled chloroplast genome is 775.90× (Supplementary Figure 3). The complete chloroplast genome of O. basilicum var. basilicum with supportive annotations was submitted to GenBank under the accession number OQ706275.1. The raw reads were deposited in the GenBank Sequence Read Archive (accession no. SRR23991168).
Figure 2. Circular map of the chloroplast genome of O. basilicum var. basilicum. From the center going outward, the first circle shows the distribution of the repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle displays the tandem repeats marked with short bars. The third circle shows the LSC, SSC, IRa, and IRb regions. The fourth circle shows the percent of GC content. The next circle shows the genes having different colors based on the functional groups. The functional classification is shown at the bottom left. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counter-clockwise direction.
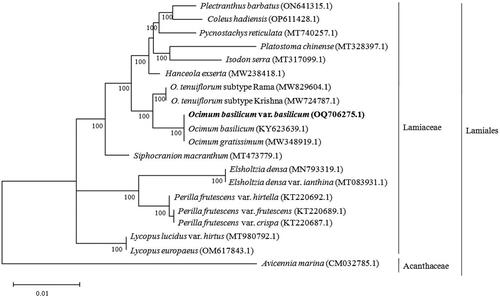
The complete chloroplast genome sequences of 19 species were retrieved from the National Center for Biotechnology Information (NCBI) to conduct the phylogenetic analysis. The complete chloroplast genome sequences were subjected to multiple sequence alignments using the MAFFT tool (Katoh and Standley Citation2013). The phylogenetic tree was constructed using MEGA software (Tamura et al. Citation2021). The tree included the chloroplast genomes of 20 species within the Lamiales order, including O. basilicum var. basilicum from this study, 18 species from the Lamiaceae family, and Avicennia marina from the Acanthaceae as an outgroup. The complete chloroplast genome sequences were utilized to understand the phylogenetic relationship among the species (). All the species of Ocimum formed a clade, and this group was closely related to Hanceola exserta. Chloroplast genome-based analysis has yielded a more robust species resolution within the Lamiaceae family, surpassing the resolution achieved through morphological characters (Sobti and Pushpangadan Citation1979) and RAPD markers (Singh et al. Citation2004). Within the Ocimum genus, our findings indicate a close relationship between O. basilicum var. basilicum and O. gratissimum, as well as O. tenuiflorum, which aligns with prior research using chloroplast DNA barcoding markers such as rbcL, matK, and psbA-trnH (Christina and Annamalai Citation2014). The complete chloroplast genome of O. basilicum var. basilicum described in detail in this study can be subsequently used for phylogenetic analysis, DNA barcoding, and molecular marker studies for the differentiation at the O. basilicum at variety level.
Figure 3. Phylogenetic tree constructed by maximum-likelihood (ML) analysis based on complete chloroplast genome sequences, including O. basilicum var. basilicum (OQ706275.1) sequenced in this study. The numbers on the nodes indicate bootstrap values with 1000 replicates. The sequences used for tree construction are as follows: Ocimum basilicum (KY623639.1; Rabah et al. Citation2017), O. gratissimum (MW348919.1; Balaji et al. Citation2021), O. tenuiflorum subtype Rama (MW829604.1; Harini et al. Citation2021), O. tenuiflorum subtype Krishna (MW724787.1; Kavya et al. Citation2021), Platostoma chinense (MT328397.1), Pycnostachys reticulata (MT740257.1; Wu et al. Citation2021), Plectranthus barbatus (ON641315.1), Coleus hadiensis (OP611428.1), Hanceola exserta (MW238418.1; Zhu et al. Citation2023), Isodon serra (MT317099.1; Zhang et al. Citation2020), Siphocranion macranthum (MT473779.1; Zhao et al. Citation2021), Lycopus lucidus var. hirtus (MT980792.1; Wang et al. Citation2021), Lycopus europaeus (OM617843.1), Elsholtzia densa (MN793319.1; Fu et al. Citation2020), Elsholtzia densa var. ianthina (MT083931.1; Yang et al. Citation2020), Perilla frutescens var. hirtella (KT220692.1), P. frutescens var. frutescens (KT220689.1), P. frutescens var. crispa (KT220687.1), and Avicennia marina (CM032785.1; Natarajan et al. Citation2021).
Conclusions
In this study, the chloroplast genome sequence of O. basilicum var. basilicum was sequenced and annotated. The phylogenetic analysis showed that the chloroplast genome of O. basilicum var. basilicum is closely related to O. gratissimum and other Ocimum species within the Lamiaceae family. This study provides valuable chloroplast genome resources of the genus Ocimum, which lay the foundation for the study of phylogenetic analysis and molecular markers to confirm the authenticity of sweet basil (O. basilicum) and its herbal products.
Author contributions
S.I. Kirankumar, R. Balaji, and Tanuja collected the specimen material, conducted the experiment, analyzed the sequence data, and drafted the paper. M. Parani contributed to the conception and design of this work. All the authors carefully read, revised, and approved the final manuscript to be published. We thank Dr. D. Narasimhan for providing his comments and suggestions on the final manuscript and Dr. Senthilkumar Umapathy for his help in the authentication of plant material.
Ethical approval
No permissions were required for the sample collection of O. basilicum var. basilicum, because it is widely distributed in the wastelands and roadsides in the tropical regions. The plant species was collected from Pallavaram, Chengalpattu District, Tamil Nadu, India (GPS coordinates: 12°58′01.4″N 80°08′42.3″E).
Supplemental Material
Download TIFF Image (3.7 MB)Supplemental Material
Download TIFF Image (13.3 MB)Supplemental Material
Download TIFF Image (9.1 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/). The complete chloroplast genome of Ocimum basilicum var. basilicum was deposited in GenBank under the accession OQ706275 (https://www.ncbi.nlm.nih.gov/nuccore/OQ706275). The associated NGS sequencing data files are available from the BioProject, Bio-Sample, and SRA submission under the accession numbers PRJNA949446, SAMN33944295, and SRR23991168, respectively.
Additional information
Funding
References
- Aminian AR, Mohebbati R, Boskabady MH. 2021. The effect of Ocimum basilicum L. and its main ingredients on respiratory disorders: an experimental, preclinical, and clinical review. Front Pharmacol. 12:805391. doi: 10.3389/fphar.2021.805391.
- Balaji R, Parani M. 2022. DNA barcoding of the market samples of single‐drug herbal powders reveals adulteration with taxonomically unrelated plant species. Diversity. 14(6):495. doi: 10.3390/d14060495.
- Balaji R, Ravichandiran K, Tanuja Parani M. 2021. The complete chloroplast genome of Ocimum gratissimum from India – a medicinal plant in the Lamiaceae. Mitochondrial DNA B Resour. 6(3):948–950. doi: 10.1080/23802359.2021.1889413.
- Christina VL, Annamalai A. 2014. Nucleotide based validation of Ocimum species by evaluating three candidate barcodes of the chloroplast region. Mol Ecol Resour. 14(1):60–68. doi: 10.1111/1755-0998.12167.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fernando G, Konstantin O, José C, Luis MC, Stefan G, Sonia T, Joaquín D, Thomas FM, Ana C. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 28(20):2678–2679.
- Fu G, Liu J, Li J. 2020. The complete chloroplast genome sequence of Elsholtzia densa, a herb with volatile aroma component. Mitochondrial DNA B Resour. 5(1):595–596. doi: 10.1080/23802359.2019.1710597.
- Harini P, Balaji R, Parani M. 2021. The complete chloroplast genome of Ocimum tenuiflorum L. subtype Rama Tulsi and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(8):2224–2226. doi: 10.1080/23802359.2021.1944381.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Claude W, dePamphilis Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kavya NM, Balaji R, Tanuja Parani M, Senthilkumar P. 2021. The complete chloroplast genome of Ocimum tenuiflorum L. subtype Krishna Tulsi and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(8):2358–2360. doi: 10.1080/23802359.2021.1951133.
- Kurnia D, Putri SA, Tumilaar SG, Zainuddin A, Dharsono HDA, Amin MF. 2023. In silico study of antiviral activity of polyphenol compounds from Ocimum basilicum by molecular docking, ADMET, and drug-likeness analysis. Adv Appl Bioinform Chem. 16:37–47. doi: 10.2147/AABC.S403175.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi: 10.1093/nar/gkw413.
- Natarajan P, Murugesan AK, Govindan G, Gopalakrishnan A, Kumar R, Duraisamy P, Balaji R, Tanuja Shyamli PS, Parida AK, Parani M. 2021. A reference-grade genome identifies salt-tolerance genes from the salt-secreting mangrove species Avicennia marina. Commun Biol. 4(1):851. doi: 10.1038/s42003-021-02384-8.
- Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JSM, Jansen RK, Ruhlman TA. 2017. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome. 10(3):1–14. doi: 10.3835/plantgenome2017.03.0020.
- Ravikumar K, Begum SN, Ved DK, Bhatt JR, Goraya GS. 2018. Compendium of traded medicinal plants. Bengaluru, India: Foundation for Revitalization of Local Health Traditions (FRLHT); p. 104.
- Rawat R, Negi KS, Mehta PS, Tiwari V, Verma SK, Bisht IS. 2016. Study of six varieties of sweet basil (Ocimum basilicum L.) and their morphological variations. J Non-Timb For Prod. 23:1–6.
- Rudayni HA, Basher NS, Al-Keridis LA, Ibrahim NA, Abdelmageed E. 2021. The efficiency of ethanolic extract of Ocimum basilicum leaves and flowers against mosquito larvae. Mortality. 6(92):6–88.
- Shahrajabian MH, Sun W, Cheng Q. 2020. Chemical components and pharmacological benefits of Basil (Ocimum basilicum): a review. Int. J. Food Propert. 23(1):1961–1970. doi: 10.1080/10942912.2020.1828456.
- Singh AP, Dwivedi S, Bharti S, Srivastava A, Singh V, Khanuja SPS. 2004. Phylogenetic relationships as in Ocimum revealed by RAPD markers. Euphytica. 136(1):11–20. doi: 10.1023/B:EUPH.0000019497.89760.e8.
- Sobti SN, Pushpangadan P. 1979. Cytotaxonomical studies in the genus Ocimum. In: Bir SS, editor. Taxonomy, cytogenetics and cytotaxonomy of plants. New Delhi: Kalyani Publication; p. 373–377.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wang Y, Wang H, Zhou B, Yue Z. 2021. The complete chloroplast genomes of Lycopus lucidus and Agastache rugosa, two herbal species in tribe Mentheae of Lamiaceae family. Mitochondrial DNA B Resour. 6(1):89–90. doi: 10.1080/23802359.2020.1847617.
- Wu P, Xu C, Chen H, Yang J, Zhang X, Zhou S. 2021. NOVOWrap: an automated solution for plastid genome assembly and structure standardization. Mol Ecol Resour. 21(6):2177–2186. doi: 10.1111/1755-0998.13410.
- Yang Y-C, Zhao Y-L, Fan Y-J. 2020. The complete chloroplast genome of Elsholtzia densa var. ianthina (Lamiaceae). Mitochondrial DNA B Resour. 5(2):1671–1672. doi: 10.1080/23802359.2020.1748533.
- Zhang HY, Ma WZ, Yan HF, Wang DQ. 2020. Characterization of the complete plastid genome of Chinese medicinal plant Isodon serra (Lamiaceae). Mitochondrial DNA B Resour. 5(3):2111–2112. doi: 10.1080/23802359.2020.1765429.
- Zhao F, Chen YP, Salmaki Y, Drew BT, Wilson TC, Scheen AC, Celep F, Bräuchler C, Bendiksby M, Wang Q, et al. 2021. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 19(1):2. doi: 10.1186/s12915-020-00931-z.
- Zhu Y, Zhang X, Yan S, Feng C, Wang D, Yang W, Daud MK, Xiang J, Mei L. 2023. SSR identification and phylogenetic analysis in four plant species based on complete chloroplast genome sequences. Plasmid. 125:102670. doi: 10.1016/j.plasmid.2023.102670.

