Abstract
In this study, the complete mitochondrial genome of Chlorogomphus shanicus Wilson, Citation2002 was reported, and the maximum-likelihood (ML) phylogenetic tree was constructed using 13 protein-coding genes (PCGs). The total length of the mitogenome of C. shanicus was 15,497 bp. Twelve PCGs started with ATN codons, except cox1 began with TTG codon. Most transfer RNA genes (tRNAs) were predicted to fold in a typical cloverleaf structure, except the trnS1 (gct), which lacked a dihydrouridine arm that had been simplified to a loop. The phylogenetic tree showed that Anisoptera was split into two clades, and revealed that C. shanicus was closely related to Cordulegaster boltonii (Donovan, 1807) which is endemic to Europe.
Introduction
South China is one of the distribution centers of dragonfly diversity in the world and hosts many endemic species. Chlorogomphus shanicus Wilson, Citation2002 (Wilson Citation2002) is an endemic species in the region. The species is recorded in Guangdong and Hunan provinces and narrowly distributed around the Nanling Mountains (preferring forests from 500 to 1000 m) (Zhang Citation2019). Chlorogomphus is a large genus of Chlorogomphidae, which was once classified into Cordulegastridae but afterward separated into Chlorogomphidae (Carle Citation1995). High potential of dispersal and a narrow species range make C. shanicus be ideal species for the study of endemicity. However, the species is very limitedly studied and no genetic information is available. Here, we reported the complete mitogenome of the C. shanicus, and performed an exhaustive phylogenetic analysis to provide further molecular information for future research on Chlorogomphidae.
Materials and methods
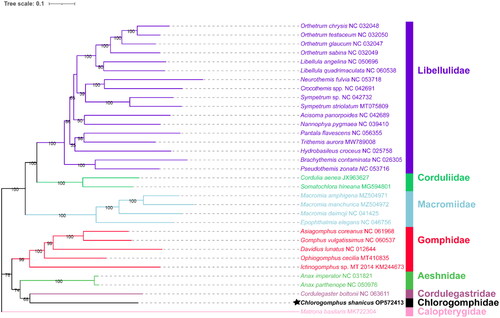
The larval specimen () was collected from Chuanlongxia Stream of Nankunshan Provincial Nature Reserve, Guangdong Province, China (23°38′47N, 113°50′36E) on 8 July 2021, and deposited in the Institute of Hydrobiology, Jinan University, Guangzhou, China under the voucher number NKS7 (https://hydrobio.jnu.edu.cn/1459/list.htm, Ningning Liu, [email protected]). Genomic DNA of the specimen was extracted from muscle tissues by a genomic DNA extraction kit (Sangon, Shanghai, China), and applied to a Next-Generation Sequencing (NGS) library construction for Illumina sequencing. We got 10x genome data, a total of 15 Gb raw data for assembly. MitoZ 2.4 (Meng et al. Citation2019) was used for mitogenome assembly, and the MITOS web server (Bernt et al. Citation2013) was used to annotate the mitogenome and predict the secondary structure of transfer RNA (tRNA). Thirty-one Anisoptera species and one outgroup (Zygoptera: Calopterygidae, Matrona basilaris Selys, 1853) were downloaded from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov). Thirteen protein-coding genes (PCGs) were concentrated to construct the phylogenetic tree. Finally, a maximum-likelihood phylogenetic tree was constructed using IQ-TREE (Nguyen et al. Citation2015) under an Edge-linked partition model for 100,000 ultrafast (Minh et al. Citation2013) bootstraps, as well as the Shimodaira–Hasegawa-like approximate likelihood-ratio test (Guindon et al. Citation2010) by PhyloSuite v1.2.2 (Zhang et al. Citation2020) ().
Figure 1. A larval specimen of C. shanicus collected at Chuanlongxia Streams of Nankunshan Provincial Nature Reserve, Guangdong Province, China. The photo was taken by Jian Liao using a Canon SLR 500D.

Table 1. Mitochondrial genomes for C. shanicus, 31 Anisoptera species, and one outgroup (M. basilaris) presented in NCBI.
Results and discussion
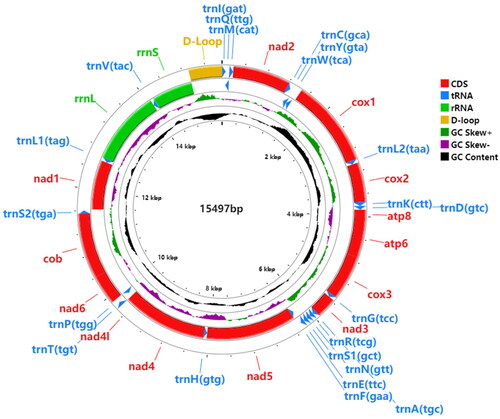
The complete mitogenome of C. shanicus (GenBank accession number: OP572413) contains 13 PCGs, 22 tRNA genes, two ribosomal RNA genes (rRNAs), and a control region (D-loop), with a total length of 15,497 bp (). Except cox1 beginning with TTG codon, 12 PCGs started with ATN codons (nad2, nad3, and nad5 with ATT, cox2, atp6, cox3, nad4L, and nad1 with ATG, atp8, and nad6 with ATC), nine PCGs terminated with a complete codon (atp8, atp6, nad4L, nad1, nad2, nad6, and cob with TAA, nad3, and nad4 with TAG); however, cox1, cox3, nad5, and nad6 ended with an incomplete codon (cox1, cox2, and nad5 with T, cox3 with TA). tRNAs had lengths from 65 bp to 72 bp, and could be folded in a typical cloverleaf structure, except the trnS1 (gct), which lacked a dihydrouridine arm that had been simplified to a loop. The 12S rRNA (rrnS) was 776 bp in length, and 16S rRNA was 1421 bp.
Figure 2. Representation of C. shanicus mitogenome: the total length was 15,497 bp, which was divided into 37 genes, including 13 PCGs, 22 tRNAs, two rRNA, and D-loop.
The phylogenetic tree () was consistent with morphological classification. It showed that Anisoptera was split into two clades, and C. shanicus was shown as a sister taxon to Cordulegaster boltonii, suggesting a close phylogenetic relatedness between Cordulegastridae and Chlorogomphidae. C. boltonii is endemic to Europe, with the only populations outside in the north of Morocco and Algeria. The two species are endemic to two different continents but close in phylogeny, suggesting that further investigation is necessary. Phylogenetic trees revealed that all families are monophyletic groups.
Conclusions
In this study, we reported the complete mitogenome of C. shanicus, which was 15,497 bp in length, containing 13 PCGs, 22 tRNA, two rRNA, and one D-loop region as a typical mitogenome. The phylogenetic tree we constructed showed Anisoptera was split into two clades, and C. shanicus was a sister taxon to C. boltonii, indicating that Chlorogomphidae was more closely related to Cordulegastridae, which was consistent with morphological classification. The phylogenetic tree revealed that all families are monophyletic groups. Our works provided the phylogenetic information of Chlorogomphidae at the mitochondrial genome level for inferring the phylogenetic relationship of Anisoptera species.
Author contributions
Haojie Wang: conceptualization, fieldwork, analysis, and writing-original draft. Lu Wang: methodology, analysis, and writing-review. Jian Liao and Bo-Ping Han: conceptualization, supervision, and writing-review and editing. All authors approve the published version of the manuscript, and agree to be accountable for all aspects of the work.
Ethical approval
This study includes no endangered animals and does not require ethical approval to collect samples.
Acknowledgements
We sincerely thank Dr. Henri J. Dumont and Dr. Eric Zeus Rizo for their valuable reading and comments. And we thank Zhenqi Wu, Jinwei Liang, and Jiancheng Liang for their help in field work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that supported the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP572413. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA886846, SRR21799098, and SAMN31143302, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Cao L, Hou W, Hu C. 2019. The complete mitochondrial genome sequence of Acisoma panorpoides Rambur, 1842 (Odonate: Libellulidae). Mitochondrial DNA B Resour. 4(2):3644–3645. doi: 10.1080/23802359.2019.1678416.
- Carle FL. 1995. Evolution, taxonomy and biogeography of ancient Gondwanian libelluloides, with comments on anisopteroid evolution and phylogenetic systematics (Anisoptera: Libelluloidea). Odonatologica. 24(4):383–424.
- Feindt W, Osigus HJ, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA B Resour. 1(1):497–499. doi: 10.1080/23802359.2016.1192504.
- Feng RQ, Luo FZ, Zhang LJ, Li M, Cao Y, Yuan ML. 2020. The complete mitochondrial genome of Sympetrum striolatum (Odonata: Libellulidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 5(2):1677–1678. doi: 10.1080/23802359.2020.1745103.
- Guan DL, Qian ZQ, Ma LB, Bai Y, Xu SQ. 2019. Different mitogenomic codon usage patterns between damselflies and dragonflies and nine complete mitogenomes for odonates. Sci Rep. 9(1):678. doi: 10.1038/s41598-018-35760-2.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi: 10.1093/sysbio/syq010.
- Jeong SY, Kim MJ, Wang AR, Kim SS, An J, Kim I. 2018. Complete mitochondrial genome sequence of the tiny dragonfly, Nannophya pygmaea (Odonata: Libellulidae). Conserv Genet Resour. 10(3):355–358. doi: 10.1007/s12686-017-0823-0.
- Kim MJ, Jeong SY, Wang AR, An J, Kim I. 2018. Complete mitochondrial genome sequence of Macromia daimoji Okumura, 1949 (Odonata: Macromiidae). Mitochondrial DNA B Resour. 3(1):365–367. doi: 10.1080/23802359.2018.1450683.
- Lan DY, Shen SQ, Cai YY, Wang J, Zhang JY, Storey KB, Yu DN. 2019. The characteristics and phylogenetic relationship of two complete mitochondrial genomes of Matrona basilaris (Odonata: Zygoptera: Calopterygidae). Mitochondrial DNA B Resour. 4(1):1745–1747. doi: 10.1080/23802359.2019.1610104.
- Ma F, Hu Y, Wu J. 2020. The complete mitochondrial genome of Anax parthenope (Odonata: Anisoptera) assembled from next-generation sequencing data. Mitochondrial DNA B Resour. 5(3):2823–2824.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for ANIMAL mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi: 10.1093/nar/gkz173.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. doi: 10.1093/molbev/mst024.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Ožana S, Dolný A, Pánek T. 2022. Nuclear copies of mitochondrial DNA as a potential problem for phylogenetic and population genetic studies of Odonata. Syst Entomol. 47(4):591–602. doi: 10.1111/syen.12550.
- Park JS, Kim MJ, Kim SS, Kim I. 2022. Complete mitochondrial genome of Asiagomphus coreanus (Odonata: Gomphidae), which is endemic to South Korea. Mitochondrial DNA B Resour. 7(5):791–793. doi: 10.1080/23802359.2022.2072246.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166. doi: 10.1093/nar/gku917.
- Wang JD, Wang CY, Zhao M, He Z, Feng Y. 2019. The complete mitochondrial genome of an edible aquatic insect Epophthalmia elegans (Odonata: Corduliidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 4(1):1381–1382. doi: 10.1080/23802359.2019.1598303.
- Wilson KDP. 2002. Notes on Chlorogomphidae from southern China, with descriptions of two new species (Anisoptera). Odonatologica. 31(1):65–72.
- Yong HS, Song SL, Suana IW, Eamsobhana P, Lim PE. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Syst Ecol. 69:124–131. doi: 10.1016/j.bse.2016.09.002.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2014. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2272–2273.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
- Zhang HM. 2019. Dragonflies and damselflies of China. Vol. 7. Chongqing, China: Chongqing University Press; p. 1003–1004.