Abstract
Ranunculus ternatus Thunb. 1784 is a plant with important medicinal values. Here we report its chloroplast genome. This chloroplast genome was 156,003 bp in length with a GC content of 37.86%. It is composed of a large single copy (LSC) of 85,397 bp and a small single copy (SSC) of 19,856 bp, which are separated by a pair of inverted repeats (IR) of 25,375 bp each. The chloroplast genome contained 110 unique genes, including 77 protein-coding genes, 4 rRNA genes, and 29 tRNA genes. Phylogenetic analysis indicated that R. ternatus was closely related to R. cassubicifolius. This chloroplast genome not only enriches the genome information of Ranunculus but also will be useful for the evolution study of the family Ranunculaceae.
Background
Ranunculus Linaeus 1753 is the largest genus in the family Ranunculaceae with a high species diversity of about 600 species widely distributed worldwide. Most species are found in the temperate to boreal regions and are less common in the tropics (Hörandl et al. Citation2005). Species classification studies have shown that Ranunculus is monophyletic in origin and is divided into two subgenera and 17 series (Tamura Citation1993). Ranunculus ternatus Thunb. 1784 is an annual herbaceous plant of the genus Ranunculus mainly distributed in the Henan and Anhui regions of China. Its roots contain a variety of lactones (Guo et al. Citation1995; Xiong et al. Citation2008a), flavonoids (Xiong et al. Citation2008b), triterpenoids (Zhao et al. Citation2008), glycosides (Tian et al. Citation2005) and two alkaloids (Zhang et al. Citation2007), which are used to treat pharyngitis, pulmonary tuberculosis, cervical scrofula, breast cancer, renal fibrosis and other diseases (Zhan et al. Citation2013; Feng et al. Citation2017; Xu et al. Citation2022).
In recent years, the chloroplast genomes of some Ranunculus species have been published, such as R. austro-oreganus (Zeng et al. Citation2021), R. cassubicifolius (Karbstein et al. Citation2022), R. flammula (Marcel et al. Citation2017), and R. japonicus (Zeng et al. Citation2021), R. macranthus (Raubeson et al. Citation2007), R. occidentalis (Zeng et al. Citation2021), R. pekinensis (Liu et al. Citation2022), R. repens (Marcel et al. Citation2017), R. reptans (Marcel et al. Citation2017), and R. sceleratus (Kim et al. Citation2023), R. yunnanensis (Rao et al. Citation2022). However, the chloroplast genome of R. ternatus has not been reported yet. Therefore, we assembled the chloroplast genome of R. ternatus and performed comparative genomic analyses with other Ranunculus species. Our primary objectives were to study the chloroplast genome of R. ternatus and to determine its phylogenetic position.
Materials and methods
Plant material, DNA extraction and sequencing
Dry leaves of R. ternatus were collected in this study from Gushi County, Xinyang City, Henan Province, China (, 115.672500°E, 31.911667°N). Our experimental studies, including the collection of plant material, were in accordance with institutional, national, or international guidelines. The sample was deposited at the herbarium of the College of Pharmaceutical Engineering, Xinyang Agriculture and Forestry University (voucher number: RT001, Min Song, [email protected]). Total genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987). The DNA library of next generation sequencing with an insert size of 300 bp was constructed and sequenced using the MGISEQ-2000 platform (Benagen Technology Company, Wuhan, Hubei), yielding ∼4 Gb of raw data, and low-quality sequences were removed to obtain clean data.
Figure 1. Species reference map of R. ternatus. This picture was taken by Min Song from the Xinyang Agriculture and Forestry University, Xinyang City, Henan Province, China. (voucher number: RT001; 113.82350°E, 31.96808°N). core features: R. ternatus is an annual herb. Root tubers clustered, tips stiff, resembling cat’s claws. Leaf blade glabrous. Flowers yellow or late white. Achenes ovoid. The tuberous roots are used medicinally for lymphatic tuberculosis.

Genome assembly and annotation
De novo genome assembly from the clean data was performed using GetOrganelle v.1.7.5 (Jin et al. Citation2020). The parameters applied for the plastome were “-R 15 -k 21,45,65,85,105 -F embplant_pt.” Samtools v.1.7 (Li et al. Citation2009) and bedtools v.2.28 (Quinlan and Hall Citation2010) were used for depth detection. The chloroplast genome was annotated using CPGAVAS2 (Shi et al. Citation2019), and PGA (Qu et al. Citation2019) with a reference genome (R. membranaceus, GenBank: NC_065303). GB2sequin (https://chlorobox.mpimp-golm.mpg.de/GenBank2Sequin.html) was then used to confirm the annotation results (Tillich et al. Citation2017). CPGView (Liu et al. Citation2023) was used to visualize the maps of the chloroplast genome, cis- and trans-splicing genes. In addition, we also performed genomic hotspot analysis using mVISTA (Frazer et al. Citation2004).
Repeat and IR boundary analysis
Simple sequence repeats (SSRs) were identified using the MISA software (Beier et al. Citation2017), including mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides with minimum numbers of 10, 5, 4, 3, 3, and 3, respectively. Additionally, REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) was used to calculate palindromic, forward, reverse, and complementary repeats with the following settings: minimum repeat size of 30 bp and hamming of 3 (Kurtz et al. Citation2001). In addition, comparisons between the boundaries of three regions (large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRs)) were generated using IRscope (Amiryousefi et al. Citation2018).
Phylogenetic analysis
We performed a phylogenetic analysis of the chloroplast genome of R. ternatus along with 15 other Ranunculus species. We extracted 77 common protein-coding genes (PCGs) from the genome annotation files using PhyloSuite v. 1.2.3 (Zhang et al. Citation2020). 77 genes were aligned using MAFFT v. 7.4 (Katoh and Standley Citation2013), and these aligned genes were concatenated. Based on the concatenation matrix, a phylogenetic tree was constructed using the maximum likelihood (ML) method implemented in IQ-TREE v. 2.1.2 (Nguyen et al. Citation2015), and the best model (GTR + F+R2) was inferred using ModleFinder (Kalyaanamoorthy et al. Citation2017). The bootstrap replicates were set to 1000. Tree visualization was performed in Figtree v. 1.4.3 (https://github.com/rambaut/figtree/releases/tag/v1.4.3).
Results
General features of the chloroplast genome
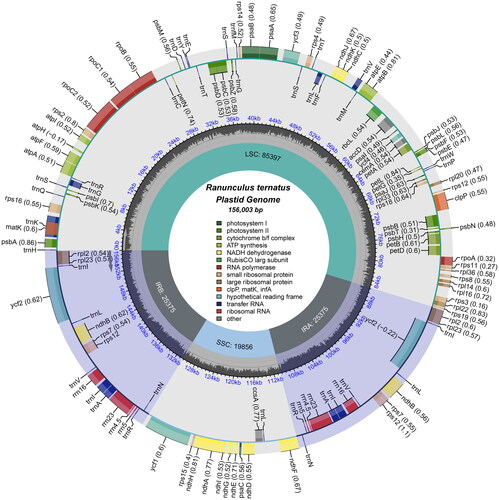
We analyzed the coverage depth of the chloroplast genome and the annotation of some difficult genes, the results indicated that the chloroplast genome of R. ternatus was similar to that of other Ranunculus species with some confidence (Figure S1; Figure S2). This chloroplast genome had a circular quadripartite structure of 156,003 bp in length (, Table S1), which consisted of a large single-copy (LSC) region (85,397 bp), a small single-copy (SSC) region (19,856 bp), and a pair of inverted repeats (IR) (25,375 bp). This chloroplast genome had a total GC content of 37.86%, the GC content in the IR region (43.46%) was significantly higher than that in the LSC region (36.03%) and the SSC region (31.30%). In addition, the annotation results showed that there were 127 genes in the chloroplast genome, leaving 110 unique genes after removing 17 duplicated genes. These unique genes included 77 PCGs, 4 ribosomal RNA genes and 29 transfer RNA genes (Table S1, Table S2). Similar to other Ranunculus species, these gene with two copies in R. ternatus contained six PCGs (ndhB, rpl2, rpl23, rps7, rps12, ycf2), seven tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) and four rRNAs (rrn4.5, rrn5, rrn16, rrn23). The mVISTA analysis showed that the IR region was more conserved than the LSC and SSC regions (Figure S3).
Figure 2. The chloroplast genome map of R. ternatus. Genes on the inside of the circle are transcribed in a clockwise direction and genes on the outside of the circle are transcribed in a counter-clockwise direction. The optional codon usage bias is displayed in the parenthesis after the gene name. The small grey bar graphs inner circle shows the GC contents. Genes are color-coded by their functional classification. The functional classification of the genes is shown in the center.
Repeat analysis
We identified 108 simple sequence repeats (SSRs) in the chloroplast genome of R. ternatus, including 31 mononucleotides, 37 dinucleotides, 17 trinucleotides, 21 tetranucleotides, one pentanucleotide, and one hexanucleotide (Table S1). Mononucleotides and dinucleotides accounted for most of the SSRs, accounting for 62.96% of the total. The SSRs were highest in the LSC region and lowest in the IR region, and were mainly concentrated in the non-coding regions (Table S3). In addition, we identified 33 long repeats, including 10 forward repeats, 5 reverse repeats, 17 palindromic repeats, and one complement repeat (Table S1), which were mainly located in the IR and LSC regions (Table S4).
IR boundaries analysis
The chloroplast genome structures of the Ranunculus species were relatively similar, with the main differences being in the location of genes within the SSC region and at the IR boundary. The ndhF genes were located within the SSC region, and the ycf1 genes were predominantly present either within the SSC region or at the IR boundary (Figure S4). In clade A, ycf1 genes were predominantly located on the IR boundary, whereas in clade B they were located within the SSC region. Compared to R. cassubicifolius, the IR region of the chloroplast genome of R. ternatus showed a small expansion, but the difference was not significant.
Phylogenetic analysis
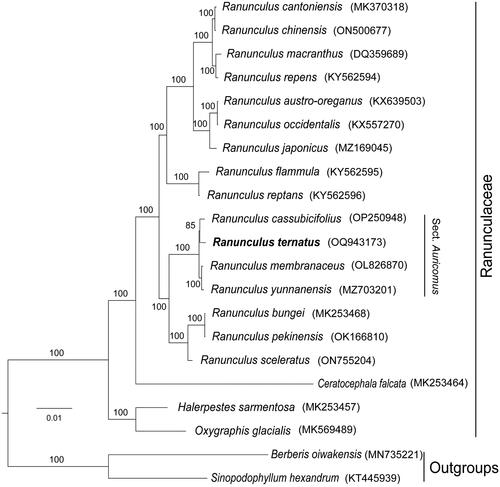
To clarify the phylogenetic position of R. ternatus in the genus Ranunculus, we performed a phylogenetic analysis. The phylogenetic tree showed that our phylogenetic results were generally consistent with previous studies, where most nodes have high bootstrap values (). In section Auricomus, the R. ternatus was more closely related to R. cassubicifolius.
Figure 3. Phylogenetic tree based on the concatenated sequences of 77 protein-coding genes in 21 species by maximum-likelihood (ML). Values split by backslashes above branches represent ML bootstraps. The best-fit model was GTR + F+R2. Branch supports were tested using ultrafast bootstrap (UFBoot) with 1000 replicates. Ranunculus ternatus (OQ943173) was marked in bold. The following sequences were used: Ranunculus cantoniensis MK370318 (Li et al. Citation2019), Ranunculus chinensis ON500677, Ranunculus macranthus DQ359689 (Raubeson et al. Citation2007), Ranunculus repens KY562594 (Marcel et al. Citation2017), Ranunculus austro-oreganus KX639503, Ranunculus occidentalis KX557270, Ranunculus japonicus MZ169045 (Zeng et al. Citation2021), Ranunculus flammula KY562595 (Marcel et al. Citation2017), Ranunculus reptans KY562596 (Marcel et al. Citation2017), Ranunculus cassubicifolius OP250948 (Karbstein et al. Citation2022), Ranunculus membranaceus OL826870, Ranunculus yunnanensis MZ703201 (Rao et al. Citation2022), Ranunculus bungei MK253468 (He et al. Citation2019), Ranunculus pekinensis OK166810 (Liu et al. Citation2022), Ranunculus sceleratus ON755204 (Kim et al. Citation2023), Ceratocephala falcata MK253464 (He et al. Citation2019), Halerpestes sarmentosa MK253457 (He et al. Citation2019), Oxygraphis glacialis MK569489 (Zhai et al. Citation2019), Berberis oiwakensis MN735221 (Xiao et al. Citation2020), Sinopodophyllum hexandrum KT445939.
Discussion
Previous phylogenetic studies on the genus Ranunculus have not included R. ternatus in the sampling range. However, some researchers have suggested that R. ternatus should be included in the section Auricomus (Hörandl and Emadzade Citation2012). In this study, we observed that R. cassubicifolius belonged to the section Auricomus and was closely related to R. ternatus. Based on this phylogenetic tree, we inferred that R. ternatus was closely related to section Auricomus. Chloroplast genomes from Ranunculus species are still scarce, and accurate phylogenetic relationships still require comprehensive analyses of the nuclear and organelle genomes of more species (Górniak et al. Citation2010). Future studies on the nuclear phylogeny of this section are needed to determine the phylogenetic relationships of Ranunculus and species within the genus. Therefore, extensive sampling of more Ranunculus species with genome sequencing and analysis will be needed in the future to resolve the complex phylogenetic relationships within Ranunculus.
Conclusion
In this study, the chloroplast genome of R. ternatus was assembled using short-read data. This chloroplast genome had a typical tetrameric structure and is similar to the chloroplast genomes of other Ranunculus plants. The phylogenetic tree supported the phylogenetic position of R. ternatus. These results clearly show that R. ternatus was located in the section Auricomus and was closely related to R. cassubicifolius. Thus, the chloroplast genome of R. ternatus not only enriches the genomic information of Ranunculus, but also lays the foundation for understanding the evolution of the genus Ranunculus.
Ethical approval
Ranunculus ternatus is widely distributed in fields and wastelands in Henan Province, and is not protected by law. Experimental research do not involve the genetic transformation, preservation of the genetic background of the species used, or any other processes requiring ethical approval. Therefore, no special permission was required.
Authors’ contributions
Xinrong Qiao and Min Song were mainly responsible for the experimental design; Xinrong Qiao, Zexia Wang, and Wei Sun participated in the genome assembly and annotation work. Xinrong Qiao and Nailiang Zhu analyzed and interpreted the data. All authors have read and approved the final manuscript.
Supplemental Material
Download MS Excel (12.6 KB)Supplemental Material
Download MS Excel (13.3 KB)Supplemental Material
Download MS Excel (13 KB)Supplemental Material
Download MS Excel (11 KB)Supplemental Material
Download JPEG Image (1.2 MB)Supplemental Material
Download JPEG Image (2.5 MB)Supplemental Material
Download JPEG Image (395.4 KB)Supplemental Material
Download JPEG Image (257.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank of NCBI (http://www.ncbi.nlm.nih.gov/) under the accession no. OQ943173. The associated BioProject, BioSample, and SRA numbers are PRJNA995920, SAMN36510160, and SRR25317840, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031. doi: 10.1093/bioinformatics/bty220.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M., 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. doi: 10.1093/bioinformatics/btx198.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Feng Z-M, Zhan Z-L, Yang Y-N, Jiang J-S, Zhang P-C., 2017. New heterocyclic compounds from Ranunculus ternatus Thunb. Bioorg Chem. 74:10–14. doi: 10.1016/j.bioorg.2017.07.004.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–279. doi: 10.1093/nar/gkh458.
- Górniak M, Paun O, Chase MW. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 56(2):784–795. doi: 10.1016/j.ympev.2010.03.003.
- Guo XM, Zhou ZL, Hong YF. 1995. Studies on the chemical constituents of Ranunculus ternatus Thunb. Acta Pharm. Sin. 30:931–934.
- He J, Yao M, Lyu R-D, Lin L-L, Liu H-J, Pei L-Y, Yan S-X, Xie L, Cheng J., 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):15285. doi: 10.1038/s41598-019-51601-2.
- Hörandl E, Paun O, Johansson JT, Lehnebach C, Armstrong T, Chen L, Lockhart P., 2005. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Mol Phylogenet Evol. 36(2):305–327. doi: 10.1016/j.ympev.2005.02.009.
- Hörandl E, Emadzade K. 2012. Evolutionary classification: a case study on the diverse plant genus Ranunculus L. (Ranunculaceae). Perspect Plant Ecol Evol Syst. 14(4):310–324. doi: 10.1016/j.ppees.2012.04.001.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Karbstein K, Tomasello S, Hodač L, Wagner N, Marinček P, Barke BH, Paetzold C, Hörandl E., 2022. Untying Gordian knots: unraveling reticulate polyploid plant evolution by genomic data using the large Ranunculus auricomus species complex. New Phytol. 235(5):2081–2098. doi: 10.1111/nph.18284.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim K-R, Park SY, Kim H, Hong JM, Kim S-Y, Yu J-N. 2023. Complete chloroplast genome determination of Ranunculus sceleratus from Republic of Korea (Ranunculaceae) and comparative chloroplast genomes of the members of the Ranunculus Genus. Genes 14(6):1149. doi: 10.3390/genes14061149.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R., 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642. doi: 10.1093/nar/29.22.4633.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.,. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352.
- Li T, Fu X, Deng H, Han X, Wen F, Xu L., 2019. The complete chloroplast genome of Ranunculus Cantoniensis. Mitochondrial DNA Part B, Resour. 4(1):1095–1096. doi: 10.1080/23802359.2019.1586483.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C., 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Liu X, Yang W, Zhang D, Liu H, Cui Y, Wang L, Hu H, Yin Y, Zhang G., 2022. The complete chloroplast genome sequence of Ranunculus pekinensis (L. Liou) Luferov (Ranunculaceae), a species endemic to China. Mitochondrial DNA Part B Resour. 7(5):841–843. doi: 10.1080/23802359.2022.2073841.
- Marcel D, Sidonie B, Sylwia S, et al. 2017. Mutation rates in seeds and seed-banking influence substitution rates across the angiosperm phylogeny. bioRxiv, 156398.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842. doi: 10.1093/bioinformatics/btq033.
- Rao R, Zeng Z-F, Du Y, Mao C-M, Xie L, Guo X-L, Yue L-L., 2022. The complete chloroplast genome of Ranunculus yunnanensis (Ranunculaceae). Mitochondrial DNA Part B Resour. 7(1):60–61. doi: 10.1080/23802359.2021.2002211.
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 8(1):174. doi: 10.1186/1471-2164-8-174.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C., 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Tamura M. 1993. Ranunculaceae Flowering Plants Dicotyledons. In: Kubitzki, K., Rohwer, J.G., Bittrich, V., editors., The families and genera of vascular plants. Berlin Heidelberg: Springer.
- Tian JK, Sun F, Cheng YY. 2005. Two new glycosides from the roots of Ranunculus ternatus. Chin Chem Lett. 16:928–930.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S., 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Xiao Q, Feng T, Yu Y. 2020. The complete chloroplast genome of Mahonia oiwakensis (Berberidaceae), a traditional Chinese medicinal plant. Mitochondrial DNA Part B Resour. 5(1):692–694. doi: 10.1080/23802359.2020.1714500.
- Xiong Y, Deng KZ, Gao WY, et al. 2008a. Studies on chemical constituents of Ranunculus ternatus. Zhongguo Zhong Yao Za Zhi. 33(8):909–911.
- Xiong Y, Deng KZ, Guo YQ, et al. 2008b. Studies on chemical constituents of flavonoids and glycosides in Ranunculus ternatus. Chin Tradit Herb Drugs. 39:1449–1452.
- Xu W, Peng R, Chen S, Wu C, Wang X, Yu T, Jian J, Zhang N, Zuo S, Chen M, et al. 2022. Ranunculus ternatus Thunb extract attenuates renal fibrosis of diabetic nephropathy via inhibiting SMYD2. Pharm Biol. 60(1):300–307. doi: 10.1080/13880209.2022.2030759.
- Zeng W-Q, Yan H-J, Wu Y-Z, Wang R-R, Xiao X-F, Qi Z-C, Yan X-L., 2021. The complete chloroplast genome sequence of Japanese buttercup Ranunculus japonicus Thunb. Mitochondrial DNA Part B Resour. 6(11):3186–3187. doi: 10.1080/23802359.2021.1987166.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y., 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21. doi: 10.1016/j.ympev.2019.02.024.
- Zhan Z, Feng Z, Yang Y, Li L, Jiang J, Zhang P., 2013. Ternatusine A, a new pyrrole derivative with an epoxyoxepino ring from Ranunculus ternatus. Org Lett. 15(8):1970–1973. doi: 10.1021/ol400643q.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT., 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
- Zhang L, Yang Z, Tian J-K. 2007. Two new indolopyridoquinazoline alkaloidal glycosides from Ranunculus ternatus. Chem Pharm Bull. 55(8):1267–1269. doi: 10.1248/cpb.55.1267.
- Zhao Y, Ruan J-L, Wang J-H, Cong Y, Song S, Cai Y-L, Fang W, Zhou D-N., 2008. Chemical constituents of radix Ranunculus ternati. Nat Prod Res. 22(3):233–240. doi: 10.1080/14786410701590343.
