Abstract
The complete mitochondrial genome sequence of Goniurosaurus varius is 17778 bp in length (GenBank accession number OQ992199), containing 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The gene order and orientation are identical to those of other Eublepharidae species in the GenBank database. Seven protein-coding genes (COX2, COX3, ND1, ND2, ND3, ND4 and CYTB) exhibit incomplete stop codon ‘T.’ Phylogenetic analysis revealed the monophyly of Goniurosaurus and Eublepharidae and suggested that G. varius is closely related to the lineage composed of G. luii and G. liboensis. Distinct from other published Eublepharidae species, G. varius contains an extra non-coding region between tRNA-Thr and tRNA-Pro, which may be formed by gene rearrangements. The complete mitochondrial genome will be helpful for further studies on the population genetics of this species and phylogenetic analyses of Eublepharidae.
Introduction
The genus Goniurosaurus Barbour, 1908a is a group of terrestrial eublepharid geckos that comprises 25 living nominal taxa in East Asia (Uetz et al. Citation2023). Goniurosaurus varius (Qi Citation2020), which is commonly named the Nanling Leopard Gecko, is a recently described new species with variable dorsal color patterns (Qi et al. Citation2020). G. varius is currently known only from the karst environment of the Nanling National Nature Reserve, northern Guangdong Province, China (Qi et al. Citation2020). The narrow distribution implies that G. varius needs more attention in conservation and evolution research. mtDNA is the main source of markers for phylogenetic research on Goniurosaurus (Jonniaux and Kumazawa Citation2008; Grismer et al. Citation2021), and the mt-genome sequence is still absent. Therefore, we assembled and analyzed the complete mitochondrial genome of G. varius for the first time. The results would be able to provide molecular data for future studies of this species.
Materials and methods
Adult G. varius were collected from Shimentai National Nature Reserve, Yingde City, Guangdong Province, China (24.780°N, 112.811°E) on 15 July 2022 (). In lizards, toe and tail clipping is commonly used for identification and DNA analysis (Ryberg et al. Citation2013; Holmes et al. Citation2016; Citation2023). We took a 0.5 cm piece of tail tissue (stored in 95% ethanol) from G. varius for genetic analyses, and then it was released immediately after treating wounds with antiseptic. Total genomic DNA was isolated from approximately 1 mm3 of tail tip tissue using the TIANamp Genomic DNA Kit (TIANGEN Biotech) according to the manufacturer’s instructions. The other tail tip tissue was deposited at Shenyang Normal University, Shenyang, China (Yu Zhou is the contact person: [email protected]) under voucher number ZY-23042301. The sequencing library was prepared by Sangon Biotech, Shanghai, China and sequenced by the Illumina HiSeq 2500 platform. The mitochondrial genome was assembled de novo using NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) with a partial mitochondrial 16S rRNA sequence of G. varius (GenBank: MW721829) (Grismer et al. Citation2021) used as the seed sequence. The average depth of coverage was generated according to the method of Ni et al. (Citation2023). The mitochondrial genome was annotated using the MITOS Webserver (Bernt et al. Citation2013) with reference to the complete Goniurosaurus luii mitogenome (GenBank: KM455054) (Li et al. Citation2016). The mitochondrial genome map of G. varius was constructed by using Chloroplot (Zheng et al. Citation2020).
Figure 1. Species reference images of Goniurosaurus varius (Qi Citation2020). the stored tail tip tissue (voucher no. ZY-23042301) was taken from the specimen in this picture ().

All six mitogenomes from the Eublepharidae species available in GenBank were downloaded as ingroup species used for phylogenetic analysis (). Two mitogenomes from the Gekkonidae species were used as outgroups (). For each of the 13 protein-coding genes (PCGs), DNA sequences were aligned using MUSCLE v3.8.31 (Edgar Citation2004) with the default parameters set. The alignments were further refined using Gblocks 9.1b (Castresana Citation2000) with the ‘codon’ model and other default settings. All refined alignments were then concatenated into the final data set. For the concatenated data set, we manually defined three partitioning strategies: unpartitioned, three partitions (one partition for each codon position), and 13 partitions (one partition for each PCG). Comparisons of the three partitioning strategies and selection of corresponding nucleotide substitution models were conducted with the Bayesian information criterion implemented in PartitionFinder (Lanfear et al. Citation2012). The 3-partition scheme was chosen as the best-fitting partitioning strategy, and all three partitions favored the GTR + G + I model. The phylogenetic relationships within Eublepharidae species were reconstructed using maximum likelihood (ML) analysis. Two species from the sister family Gekkonidae were included as outgroups (). The ML tree was estimated by using RAxML version 8.0 (Stamatakis Citation2014) with the ‘GTRGAMMAI’ model. Support for nodes in the ML tree was assessed with a rapid bootstrap analysis (option -f a) with 100 replicates.
Figure 2. Mitochondrial genome map of G. varius. Arrows are the transcribed direction of genes. GC and at contents across the mitochondrial genome are shown with dark and light shading, respectively, inside the inner circle.
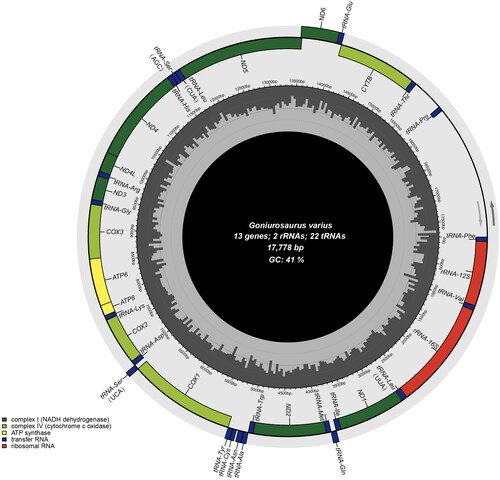
Figure 3. Maximum likelihood (ML)-based phylogenetic relationships of nine Eublepharidae species based on 13 PCGs.
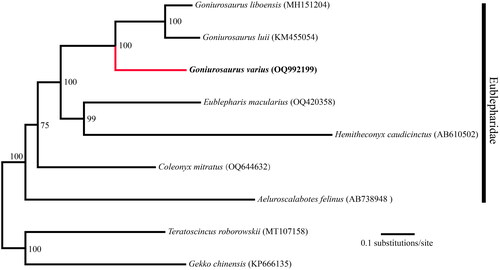
Table 1. Species and GenBank accession numbers of PCGs used in this study.
Results
The circular mitochondrial genome of G. varius was successfully assembled (OQ992199) with a length of 17,778 bp and an average coverage of 328.9 (Figure S1, supplementary material). The mitogenome of G. varius contains 13 PCGs, 22 tRNA genes, and 2 rRNA genes (). The mitochondrial gene composition and order in G. varius are identical to those in other Eublepharidae mitogenomes (Jonniaux et al. Citation2012; Li et al. Citation2016). Seven PCGs (COX2, COX3, ND1, ND2, ND3, ND4 and CYTB) use the stop codon of TAA. In contrast to other Eublepharidae species, the mitogenome of G. varius contains a 456 bp non-coding region between tRNA-Thr and tRNA-Pro (). The nucleotide composition of this mitogenome is A 32.71%, C 27.88%, G 13.39%, and T 26.01%, with an AT-biased content of 58.72%, which is similar to the 61.5% of Goniurosaurus luii (GenBank: KM455054). Phylogenetic relationships among the mitogenomes of 9 Eublepharidae species were successfully reconstructed with strong support (). Phylogenetic analysis revealed the monophyly of Goniurosaurus and Eublepharidae (). The phylogenetic tree showed that G. varius was the most closely related to the lineage that was composed of G. luii and G. liboensis.
Discussion and conclusions
The number and identity of the genes encoded in the mitochondrial (mt) genome are highly conserved in bilaterian animals, while the order and orientation and extra non-coding region in which they appear on the circular double-strand DNA molecule are not (Boore Citation1999). Gene rearrangements are known to be the main reason for mitogenome structural differentiation. In this study, G. varius contains a non-coding region between tRNA-Thr and tRNA-Pro, which is different from other published Eublepharidae species. We hypothesize that this extra non-coding region was formed by gene rearrangements, and similar formations were also observed in other animals (Sano et al. Citation2005; Kilpert et al. Citation2012; Kurabayashi and Sumida Citation2013).
The complete mitogenome of G. varius presented here will be useful for investigating the phylogeny and population genetics of this species, Goniurosaurus and even Eublepharidae.
Author contributions
YZ, PL and ZZ designed and conceived this work; LD, ZL, Longming Fu collected the samples; LD, LX and HC performed the DNA isolation; ZL, Lin Feng and SY performed the mtgenome assembly; Longming Fu, Lin Feng and SY performed the phylogenetic analysis; ZZ and YZ wrote the first version of the manuscript; and PL and ZZ carried out the investigation. All the authors have read, revised and approved the final manuscript.
Ethics statement
The Committee on the Ethics of Animal Experiments of the College of Life Sciences, Shenyang Normal University, permitted and approved the study (SYNU-20220203) to comply with the International Union for Conservation of Nature (IUCN) policies for research involving species at risk of extinction.
Supplemental Material
Download MS Word (162 KB)Acknowledgement
We want to express our gratitude to the Institute of Herpetology, Shenyang Normal University, for the facilities and support of this research project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of the NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ992199. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA995366, SRX21042629, and SAMN36468329, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Boore J. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. doi: 10.1093/nar/27.8.1767.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi: 10.1093/nar/gkh340.
- Grismer LL, Ngo HN, Qi S, Wang Y-Y, Le MD, Ziegler T. 2021. Phylogeny and evolution of habitat preference in Goniurosaurus (Squamata: Eublepharidae) and their correlation with karst and granite-stream-adapted ecomorphologies in species groups from Vietnam. Vertebr Zool. 71:335–352. doi: 10.3897/vz.71.e65969.
- Hao S, Ping J, Zhang Y. 2016. Complete mitochondrial genome of Gekko chinensis (Squamata, Gekkonidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4226–4227. doi: 10.3109/19401736.2015.1022751.
- Holmes IA, Mautz WJ, Davis Rabosky AR. 2016. Historical environment is reflected in modern population genetics and biogeography of an island endemic lizard (Xantusia riversiana reticulata). PLoS One. 11(11):e0163738. doi: 10.1371/journal.pone.0163738.
- Holmes IA, Monagan IV, Jr Westphal MF, Johnson PJ, Davis Rabosky AR. 2023. Parsing variance by marker type: testing biogeographic hypotheses and differential contribution of historical processes to population structure in a desert lizard. Mol Ecol. 32(17):4880–4897. doi: 10.1111/mec.17076.
- Jonniaux P, Hashiguchi Y, Kumazawa Y. 2012. Mitochondrial genomes of two African geckos of genus Hemitheconyx (Squamata: Eublepharidae). Mitochondrial DNA. 23(4):278–279. doi: 10.3109/19401736.2012.668898.
- Jonniaux P, Kumazawa Y. 2008. Molecular phylogenetic and dating analyses using mitochondrial DNA sequences of eyelid geckos (Squamata: Eublepharidae). Gene. 407(1–2):105–115. doi: 10.1016/j.gene.2007.09.023.
- Kilpert F, Held C, Podsiadlowski L. 2012. Multiple rearrangements in mitochondrial genomes of Isopoda and phylogenetic implications. Mol Phylogenet Evol. 64(1):106–117. doi: 10.1016/j.ympev.2012.03.013.
- Kurabayashi A, Sumida M. 2013. Afrobatrachian mitochondrial genomes: genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genomics. 14(1):633. doi: 10.1186/1471-2164-14-633.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. doi: 10.1093/molbev/mss020.
- Li HM, Hou LX, Zhang Y, Guo DN, Liu YJ, Qin XM. 2016. Complete mitochondrial genome of Goniurosaurus luii (Squamata, Eublepharidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2131–2132. doi: 10.3109/19401736.2014.982591.
- Lisachov A, Tishakova K, Romanenko S, Lisachova L, Davletshina G, Prokopov D, Kratochvíl L, Brien O, Ferguson-Smith P, Borodin M, et al. 2023. Robertsonian fusion triggers recombination suppression on sex chromosomes in Coleonyx geckos. Sci Rep. 13(1):15502. doi: 10.1038/s41598-023-39937-2.
- Ma JL, Dai XY, Xu XD, Guan JY, Zhang YP, Zhang JY, Yu DN. 2020. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: gekkonidae) and its phylogeny. Mitochondrial DNA Part B. 5(2):1575–1577. doi: 10.1080/23802359.2020.1742608.
- Ni Y, Li J, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. doi: 10.17504/protocols.io.4r3l27jkxg1y/v1.
- Pinto BJ, Gamble T, Smith CH, Keating SE, Havird JC, Chiari Y. 2023. The revised reference genome of the leopard gecko (Eublepharis macularius) provides insight into the considerations of genome phasing and assembly. J Hered. 114(5):513–520. doi: 10.1093/jhered/esad016.
- Qi S, Grismer LL, Lyu ZT, Zhang L, Li PP, Wang YY. 2020. A definition of the Goniurosaurus yingdeensis group (Squamata, Eublepharidae) with the description of a new species. Zookeys. 986:127–155. doi: 10.3897/zookeys.986.47989.
- Ryberg WA, Hill MT, Painter CW, Fitzgerald LA. 2013. Landscape pattern determines neighborhood size and structure within a lizard population. PLoS One. 8(2):e56856. doi: 10.1371/journal.pone.0056856.
- Sano N, Kurabayashi A, Fujii T, Yonekawa H, Sumida M. 2005. Complete nucleotide sequence of the mitochondrial genome of Schlegel’s tree frog Rhacophorus schlegelii (family Rhacophoridae): duplicated control regions and gene rearrangements. Genes Genet Syst. 80(3):213–224. doi: 10.1266/ggs.80.213.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Uetz P, Freed P, Aguilar R, Reyes F, Hošek J. 2023. The reptile database. [accessed 2023 Oct 10]. www.reptile-database.org.
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 11:576124. doi: 10.3389/fgene.2020.576124.
