Abstract
Impatiens huangyanensis Jin and Ding Citation2002 is a plant species with very small populations, and it distributes only in Huangyan, Zhejiang Province, China. In this study, the complete chloroplast genome of I. huangyanensis was assembled by using high-throughput Illumina paired-end sequences. Its genomic feature was determined, and comparative genomic analysis of the genus Impatiens was performed. The results revealed that the full-length chloroplast genome of I. huangyanensis was 152,156 bp with a GC content of 36.8%. The chloroplast genome contains a typical quadripartite structure, comprising two copies of inverted repeats (IRs), a small single-copy (SSC) region, and a large single-copy (LSC) region. The sequence lengths of IR, SSC, and LSC were 25,756 bp, 17,662 bp, and 82,982 bp, respectively. The chloroplast genome consisted of 134 genes, including 84 protein-coding genes, 37 transfer RNA genes, eight ribosomal RNA genes, and five pseudogenes. Phylogenic results indicated I. huangyanensis shared a clade with I. davidii Franchet 1883, I. macrovexilla Chen 2000, I. fanjingshanica Chen 1999, and I. piufanensis Hook 1908, with a support rate of 100%. Our study provided insight into further studies on the conservation genetics of I. huangyanensis.
Introduction
The Balsaminaceae is a widely distributed family that includes only two genera, namely Hydrocera and Impatiens. Hydrocera is a monospecific genus, containing only one species H. triflora, distributed in tropical Asia such as Southeast Asia, South India, and China (Shui et al. Citation2011). Impatiens is a species-rich genus, comprising over 1,000 species, mostly annual and perennial herbs with succulent stems (Janssens et al. Citation2006). China is regarded as the center for origin and diversification of Balsaminaceae, and there are about 250 wild Impatiens species, many of which have long been utilized as medicinal herbs (Luo et al. Citation2021). I. huangyanensis is a narrowly distributed Impatiens species, which was found only in mountain areas of Huangyan, Zhejiang Province, occurring in habitats of roadsides and forest margins, with very small populations (Jin and Ding Citation2002). At present, the chloroplast genome of I. huangyanensis has not been reported, and its systematic genetic location remains unclear. In this study, the chloroplast genome of I. huangyanensis was assembled based on high-throughput paired-end sequences, and a phylogenetic tree was generated to reveal its relationship with other Impatiens species.
Materials and methods
Plant sampling
Fresh leaves were gathered from Foling (28°32′25″ N, 121°09′37″ E), Huangyan, Zhejiang Province, China (). Leaves were taken to the laboratory and then washed with running water to get rid of dirt and dust before rinsed with sterile distilled water. A voucher specimen namely CHS20200388 was deposited in the Molecular Biology Innovation Laboratory at Taizhou University (Dr. Ming Jiang, [email protected]).
Figure 1. Impatiens huangyanensis Jin and Ding Citation2002. (A) Flower lateral view; (B) flower front view; (C) fruits; (D) natural habitat of I. huangyanensis. All the photos were taken by Ming Jiang.

DNA isolation, sequencing, assembling, and annotation of the chloroplast genome
The sodium dodecyl sulfate method was applied to extract high-quality genomic DNA. The genomic DNA was used to construct paired-end sequencing libraries with an average insert size of 350 bp. The library was sequenced by an Illumina Hiseq X Ten sequencing platform. Low-quality reads were filtered by using NGSQCToolkit v2.3.3 (Patel and Jain Citation2012). Chloroplast genome assembly was performed using the NOVOPlasty program (Dierckxsens et al. Citation2017). The chloroplast genome was annotated by Dual Organellar GenoMe Annotator (Wyman et al. Citation2004). Transfer RNA gene prediction was conducted by tRNAscan-SE 2.0.9 (Chan and Lowe Citation2019). The whole chloroplast genome map of I. huangyanensis was drawn using CPGView (http://www.1kmpg.cn/cpgview/; Liu et al. Citation2023). Sliding window analysis of nucleotide diversity was performed by using DnaSP 6.0 (Rozas et al. Citation2017).
Phylogenetic analysis
To understand its relationship with other Impatiens species, 18 chloroplast genome sequences of Impatiens species were downloaded from GenBank (National Center for Biotechnology Information [NCBI]) to construct a phylogenetic tree. We also downloaded a sequence of H. triflora (L.) Wight. et Arn. 1753 (NCBI accession number: NC_037400), which was used as an outgroup species. The chloroplast genomes were aligned with MAFFT v7.450, a multiple sequence alignment program (Katoh and Standley Citation2013). Based on the best model GTR + R, a phylogenetic tree was built by PhyML 3.1 with the maximum-likelihood method using whole chloroplast genome sequences (Guindon et al. Citation2010).
Results
Totally, 3.27G clean data were obtained, with 10,886,063 reads. The results revealed the complete chloroplast genome of I. huangyanensis was 152,156 bp in length, with an average depth of 3164.80× (Supplementary Figure S1). Among the Impatiens chloroplast genomes used in this study, the sequence lengths ranged from 151,538 bp (I. fanjingshanica Chen 1999) to 152,928 bp (I. mengtszeana Hooker 1908). The chloroplast genome consisted of four regions, a large single-copy (LSC), a small single-copy (SSC), and two inverted repeats (IRs), and the lengths of LSC, SSC, and IRs were 82,982 bp, 17,662 bp, and 25,756 bp, respectively (). The GC content of the plastid genome was 36.8%, which was equal to those of I. fanjingshanica, I. hawkeri Bull 1887, I. walleriana Hooker 1868, I. glandulifera Royle 1834, I. piufanensis Hook 1908, I. cyanantha Hook 1908, I. macrovexilla Chen 2000, and I. uliginosa Franchet 1886. IRs of Impatiens plant species used in this study were not conserved, they varied from 82,542 bp (I. fanjingshanica) to 83,741 bp (I. chlorosepala Handel-Mazzetti 1934). The plastid genome sequence of I. huangyanensis was submitted to GenBank with an accession number of OR139616.
Figure 2. The chloroplast genome of Impatiens huangyanensis. The map contains six tracks. From the center outward, the first track shows the dispersed repeats, which consist of direct and palindromic repeats, connected with red and green arcs. The second track indicates the long tandem repeats as short blue bars. The third track reveals the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, type of repeat they represent, and the description of the repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). the chloroplast genome contains an LSC region, an SSC region, and two IR regions, and they are shown on the fourth track. The GC content along the genome is shown on the fifth track. Genes are color-coded according to their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The bottom left corner indicates the key for the functional classification of the genes.
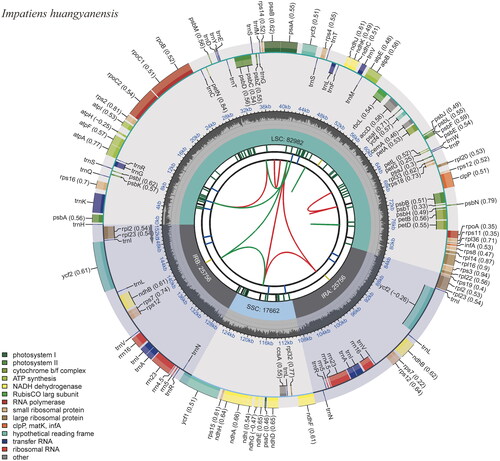
The chloroplast genome consisted of 134 genes, including 84 protein-coding genes, 37 tRNA genes, eight rRNA genes, and five pseudogenes. Eighteen genes including ndhB, rpl23, trnA-UGC, trnG-GCC, trnI-CAU, trnI-GAU, trnL-CAA, trnR-ACG, trnV-GAC, ycf1, ycf2, ycf15, rpl2, rps12, rrn4.5, rrn5, rrn16, and rrn23 contained two copies. A total of 13 genes had one or two introns, these genes included atpF, clpP, ndhA, and ndhB (two copies), petB, petD, and rpl2 (two copies), rpl16, rpoC1, rps16, and ycf3 (Supplementary Figure S2). The matK and both copies of ycf15 were found to be pseudogenized due to the presence of internal stop codons, while the ycf1 gene overlapped with ndhF on SSC/IR border and rps19 on IR/LSC border were pseudogenized owing to truncations at their 3′ ends. The rps12 is a trans-splicing gene (Supplementary Figure S3). By using DnaSP, trnG-GCC, ndhF-rpl32, and ycf1 were identified as highly variable regions (Supplementary Figure S4).
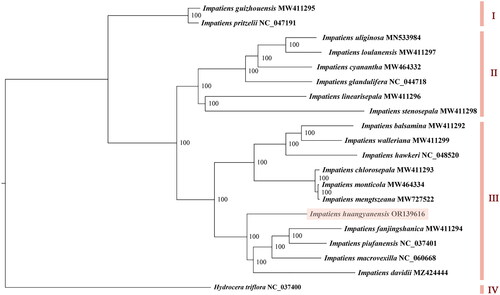
The phylogenetic analysis results showed that the 20 Balsaminaceae species clustered into four major groups. Both I. guizhouensis Chen 1999 and I. pritzelii Hook 1908 shared group I, and I. uliginosa, I. loulanensis Hooker 1911, I. cyanantha, I. glandulifera, I. linearisepala Akiyama Ohba & Wu 1996, and I. stenosepala Pritzel 1900 gathered in group II. The outgroup species H. triflora was alone in group IV, while the remaining 11 species, including I. huangyanensis, were in another group (III). I. huangyanensis was sister to I. davidii Franchet 1883, I. macrovexilla, I. fanjingshanica, and I. piufanensis, with a support rate of 100% ().
Figure 3. The maximum-likelihood tree based on complete chloroplast genome sequences of Impatiens huangyanensis and 18 other Impatiens species, with Hydrocera triflora as the outgroup species. The numbers next to the nodes show bootstrap support values. The phylogenetic tree was generated by PhyML 3.1 with the maximum-likelihood method.
Discussion and conclusions
Impatiens is a species-rich genus of angiosperms, and a number of new species were described (Tiwari Citation2023). However, the chloroplast genomes of most Impatiens species have not yet been assembled. In this study, we assembled I. huangyanensis chloroplast genome and revealed its close relationship with I. davidii, I. macrovexilla, and I. piufanensis. Pseudogenization is a common phenomenon in chloroplast genome. In our present study, five genes were found to be pseudogenized. The pseudolization of matK was also observed in Anthoceros formosae Steph. 1916, Campylotropis bonii Schindl. 1916, and some photosynthetic orchid species (Kugita et al. Citation2003; Barthet et al. Citation2015; Feng et al. Citation2022). One copy of ycf1 is located at the boundary of SSC/IR, and its 3′ ends were truncated, pseudogenization of ycf1 is common due to the incomplete duplication of the normal copy (Amar Citation2020).
Assembly and phylogenetic analysis of I. huangyanensis chloroplast genome sequence provided useful data for further studies on population structure and conservation of Impatiens genus.
Ethical approval
The authors declare that the collection of samples complies with the rules and regulations of relevant institutional, national guidelines and legislation, and they did not cause damage to the local environment.
Author contributions
Ming Jiang and Junfeng Wang were responsible for the conception and design of the study. Ming Jiang and Yan Zhu performed the data analysis. Yan Zhu, Honghua Bao, and Huijuan Zhang wrote the paper. Honghua Bao and Huijuan Zhang annotated the chloroplast genome. All the authors were involved in the analysis and interpretation of the data, revising the manuscript critically for intellectual content, and giving the final approval of the version to be published. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (567.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ OR139616. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA897866, SRR22178406, and SAMN31582508, respectively.
Additional information
Funding
References
- Amar MH. 2020. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J Genet Eng Biotechnol. 18(1):42. doi: 10.1186/s43141-020-00057-3.
- Barthet MM, Moukarzel K, Smith KN, Patel J, Hilu KW. 2015. Alternative translation initiation codons for the plastid maturase MatK: unraveling the pseudogene misconception in the Orchidaceae. BMC Evol Biol. 15(1):210. doi: 10.1186/s12862-015-0491-1.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955.
- Feng Y, Gao XF, Zhang JY, Jiang LS, Li X, Deng HN, Liao M, Xu B. 2022. Complete chloroplast genomes provide insights into evolution and phylogeny of Campylotropis (Fabaceae). Front Plant Sci. 13:895543. doi: 10.3389/fpls.2022.895543.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. doi: 10.1093/sysbio/syq010.
- Janssens S, Geuten K, Yuan YM, Song Y, Küpfer P, Smets E. 2006. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Syst Bot. 31(1):171–180. doi: 10.1600/036364406775971796.
- Jin XF, Ding BY. 2002. A new species of Impatiens from eastern Zhejiang, China. Acta Phytotax Sin. 40(2):167–169.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kugita M, Kaneko A, Yamamoto Y, Takeya Y, Matsumoto , Yoshinaga K. 2003. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucl Acid Res. 31:716–721. doi: 10.1093/nar/gkg155.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGview: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Luo C, Huang W, Sun H, Yer HY, Li XY, Li Y, Yan B, Wang Q, Wen YH, Huang MJ, et al. 2021. Comparative chloroplast genome analysis of Impatiens species (Balsaminaceae) in the karst area of China: insights into genome evolution and phylogenomic implications. BMC Genomics. 22:571. doi: 10.1186/s12864-021-07807-8.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619. doi: 10.1371/journal.pone.0030619.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302. doi: 10.1093/molbev/msx248.
- Shui YM, Janssens S, Huang SH, Chen WH, Yang ZG. 2011. Three new species of Impatiens L. from China and Vietnam: preparation of flowers and morphology of pollen and seeds. Syst Bot. 36(2):428–439. doi: 10.1600/036364411X569615.
- Tiwari UT. 2023. Impatiens tajoensis (Balsaminaceae): a new species from Arunachal Pradesh, India. Taiwania. 68(1):39–43.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. doi: 10.1093/bioinformatics/bth352.
