Abstract
In this paper, we first report the complete mitochondrial genome of Neolissochilus soroides. The main purpose of this study was to determine the mitochondrial genome and phylogenetic status of N. soroides. The length mitogenome was 16584 bp, containing 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and 3 non-coding control regions. The genome showed a slight A + T bias (A + T = 56.47%). 12 genes (ND1, COX2, ATP6, ND4L, ND5, ND6, ND2, ATP8, ND3, ND4, Cytb, COX3) start with ATG codon, besides one gene (COX1) start with GTG codon. Six genes (ND1, COX1, ATP6, ND4L, ND5, ND6) end with a TAA codon, 3 genes (ND2, ATP8, ND3) end with a TAG codon, and four genes (COX2, ND4, Cytb, COX3) end with the TA or T codon. The phylogenetic analysis showed that N. soroides was closely related to N. hendersoni. The mitogenome could have important implications for phylogeny, population genetics, and conservation of the N. soroides.
Introduction
Neolissochilus soroides (Duncker, Citation1904) is a freshwater ornamental carp that lives in fast-flowing rivers and streams with high levels of dissolved oxygen. It is mainly distributed in Southeast Asia (Rainboth, Citation1996) and had also been reported in China (Tan & Lim, Citation2004). In the past, Tor (now placed in Neolissochilus) soso was treated as the synonym name of N. soroides. Due to the loss of the holotype, the classification of T. soso requires additional material (Scharpf, Citation2015). And the lateral-line scales which are the main morphological features of N. soroides are 20–24, rarely 20; no black line on the lateral-line scales; the ventral fin has fin hooks in orange, and the scales are yellowish brown () (Khaironizam, Akaria-Ismail, & Armbruster, Citation2015). In addition to numerical analysis methods for morphological traits, molecular phylogenetic analysis, which assesses biological evolution based on biomolecular sequence differences, is increasingly being applied. Yang et al. (Citation2015) used sequences from the mitochondrial gene (Cytb) to analyze the phylogenetic relationships of N. soroides in Cypriniformes. Nevertheless, until now, the complete mitochondrial genome of N. soroides could not be retrieved in NCBI. In this paper, we first reported the complete mitochondrial genome of N. soroides, and analyzed the main structural information and the phylogenetic relationships, which could provide useful information for studying genetic diversity and phylogeny conservation.
Materials and methods
Sample collection and preservation
The fish sample of this study was collected from Daying River, Tengchong City, Yunnan Province, China (24°36′36ʺ N, 97°49′12ʺ E). The specimen was euthanized by snap-frozen in liquid nitrogen and then transferred by stored in dry ice to the laboratory of Zhejiang Marine Fisheries Research Institute (http://www.zjhys.cn, Ye Chen, [email protected]) with the voucher number HZ20211021. The N. soroides fish sample was identified by morphometric measurements and meristic counts using calipers and a dissecting microscope, based on the morphological characteristics described by Khaironizam et al. (Citation2015). Then the muscle sample from the back of the N. soroides was dissected and preserved in 95% ethanol for total genomic DNA extraction.
Mitochondrial genome sequencing
Total genomic DNA was extracted following the protocol described by Wei et al. (Citation2022). After the sample genomic DNA had been tested for sequencing compliance, the DNA was sheared off by physical means (ultrasound). Then, the interrupted DNA was purified to construct a sequencing library, and the steps were as follows: (1) DNA end repair, 3′ end addition A, sequencing junction ligation. (2) Using Agarose gelation to recover the target fragments by gel electrophoresis. (3) Target fragment amplification by PCR. (4) Building sequencing libraries. They were then sequenced by Illumina NovaSeq6000 platform (Illumina, San Diego, CA).
Assembly, annotation, and analysis
Getorganells splicing software (https://github.com/Kinggerm/GetOrganelle) was used to splice the sequenced reads in multiple iterations, and the initial assembly results were obtained. Then, the clean reads were compared back to the mitochondrial genome sequence, and the bases were corrected using Pilon v1.23 (Walker et al. Citation2014) and annotated using MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Ren et al. Citation2020). The mitochondrial circle mapping was done by OGDRAW (https://chlorobox.mpimp-golm.mpg.de/O).
GDraw.html). In order to explore the phylogenetic position of N. soroides, we used the whole mitochondrial data from the nucleotide and performed multiple sequence alignments. After multiple sequence alignments, a phylogenetic tree by MEGA 7.0 (Kumar et al. Citation2016) was generated, using maximum likelihood (ML) to performe phylogenetic analysis, and generated 1000 bootstraps. The phylogenetic trees in this study include 24 fish species belonging to Neolissochilus, Tor, Acorssocheilus, and Puntius (as an outgroup).
Results
Genomic analysis results
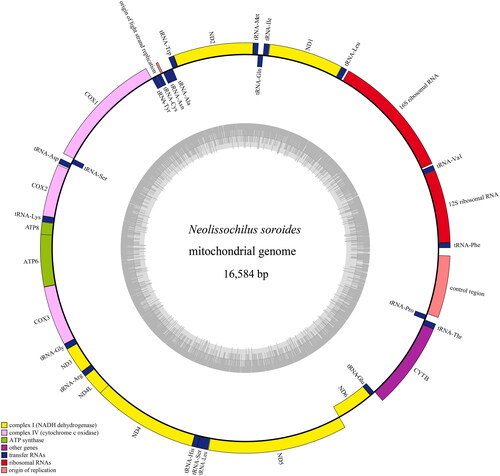
The genome of N. soroides was assembled correctly (Figure S1, supplementary material). The entire mitochondrial genome of N. soroides (GenBank accession no. OM202554) had a length of 16,584 bp. The overall base composition of the mitogenome was 31.8% for A, 27.76% for C, 15.78% for G and 24.67% for T. The percentage of A + T content was 56.47%. The gene consists of 13 protein-coding genes (COX1, COX2, ATP8, ATP6, COX3, ND3, ND1, ND5, ND4, ND4L, ND6, Cytb, ND2), two rRNA genes (12S rRNA, 16S rRNA), 22 tRNA genes and three non-coding control regions (). Most of the sequence elements were located on the heavy strand, except one protein-coding gene and eight tRNA (ND6 and Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro). The three non-coding controls were 32, 45, and 816 bp long respectively. Among 13 protein-coding genes,12 genes (ND1, COX2, ATP6, ND4L, ND5, ND6, ND2, ATP8, ND3, ND4, Cytb, COX3) were started with ATG codon, besides one gene (COX1) was started with GTG codon. And six genes (ND1, COX1, ATP6, ND4L, ND5, ND6) ended in the TAA codon, three genes (ND2, ATP8, ND3) ended in the TAG codon, and four genes (COX2, ND4, Cytb, COX3) ended in the TA or T codon.
Figure 2. Mitochondrial genome map of Neolissochilus soroides. H-strand is located in the outer ring and L-strand is located in the inner ring. The gene consists of 13 protein-coding genes, two rRNA genes, 22 tRNA genes and three non-coding control regions. Among them, yellow: complex I (NADH dehydrogenase); light green: complex III (ubiquinol cytochrome c reductase); pink: complex IV (cytochrome c oxidase); dark green: ATP synthase; blue: transfer RNAs; red: ribosomal RNAs.
Phylogenetic analysis
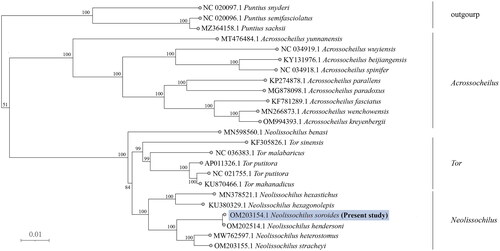
The phylogenetic position of N. soroides in subfamily Neolissochilus was shown in . soroides was closely related to N. hendersoni (OM202514.1). The phylogenetic analysis showed that N. soroides was clustered together with N. hendersoni.
Figure 3. Phylogenetic analysis of N. soroides based on the entire mtDNA genome sequences of 24 Cypriniformes available in GenBank. Numbers above the nodes indicate 1000 bootstrap values. Accession numbers are shown before species names. The following sequences were used: Neolissochilus hendersoni OM202514.1 (Guo et al. Citation2023), Neolissochilus stracheyi OM203155.1 (Wei, Z. et al. 2022), Neolissochilus heterostomus MW762597.1 (He et al. Citation2021), Neolissochilus hexagonolepis KU380329.1 (Zhou et al. Citation2016), Neolissochilus hexastichus voucher MN378521.1 (Singh et al. Citation2021), Tor sinensis KF305826.1 (Huang et al. Citation2015), Tor malabaricus NC 036383.1 (Chandhini et al. Citation2019), Tor putitora AP011326.1 (Sati et al. Citation2014), Tor mosal mahanadicus KU870466.1 (Sarma et al. Citation2022), Tor putitora NC 021755.1 (Sati, J. et al. 2014), Neolissochilus benasi MN598560.1 (Gu et al. Citation2020), Acrossocheilus yunnanensis MT476484.1 (Chen et al. Citation2022), Acrossocheilus spinifer NC 034918.1 (Zhao et al. Citation2022), Acrossocheilus beijiangensis KY131976.1 (Liu et al. Citation2018), Acrossocheilus wuyiensis NC 034919.1 (Yuan et al. Citation2017), Acrossocheilus paradoxus MG878098.1 (Ju et al. Citation2018), Acrossocheilus parallens KP274878.1 (Xie et al. Citation2016), Acrossocheilus fasciatus KF781289.1 (Cheng et al. Citation2015), Acrossocheilus kreyenbergii OM994393.1 (Zhou et al. Citation2023), Acrossocheilus wenchowensis MN266873.1 (Pan et al. Citation2019), Puntius snyderi NC 020097.1 (Jang-Liaw et al. Citation2013), Puntius sachsii MZ364158.1 (Sun et al. Citation2023), Puntius semifasciolatus NC 020096.1 (Sun et al. Citation2023), Puntius chalakkudiensis NC 018566.1 (Khare et al. Citation2014).
Discussion and conclusion
Three non-coding regions include an L-strand replication origin and two control regions, which were different from Neolissochilus heterostomus (He et al. Citation2021), and similar with Garra motuoensis (Gong et al. Citation2022).
The phylogenetics indicates that N. soroides belonged to Neolissochilus and not to Tor, which was the same as Khaironizam et al. (Citation2015). Phylogenetic tree in this paper showed that Neolissochilus and Tor and Acrossocheilus have been reasonably identified by morphology. However, N. benasi (MN598560.1) did not belong to the genus Neolissochilus, which was not consistent with the study of Gu et al. (Citation2020). In this regard, we hypothesize that the inconsistent results may be because the reliability (numbers on branches) of N. benasi phylogenetic trees in the study of Gu et al. (Citation2020) was not high enough.
The complete mitochondrial genome of N. soroides was sequenced and annotated using high-throughput sequencing technology. The basic genetic data provided in this paper could provide a reference for further research on the genetics and evolutionary study of N. soroides.
Author contributions statement
Ye Chen and Yongyao Guo designed the experiment and wrote the original manuscript. Yongyao Guo, Zhenzhu Wei and Xiaoli Zhao provided sample collection and analyzed the data. All authors approved the final manuscript and agreed to be accountable for all aspects of the study.
Ethics statement
All animal protocols have been reviewed and approved by the Ethics Review Committee for Experimental Animal Welfare of Zhejiang Ocean University. The approval number is 20210504.
Supplemental Material
Download MS Word (79.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/OM203154.1/) under accession no. OM203154. The associated BioProject, SRA, and Bio-Sample Numbers are PRJNA817098, SRR18356122, and SAMN26747790.
Additional information
Funding
References
- Chandhini S, Arjunan VM, Anvar Ali PH, Raghavan R, Pillai D, Rejish Kumar VJ. 2019. Complete mitogenome analysis of endangered Malabar mahseer (Tor malabaricus). Conservation Genet Resour. 11(2):185–189. doi: 10.1007/s12686-018-0997-0.
- Chen Q, Lian X, Li Y, Liu Z, Shen Y. 2022. Complete mitochondrial genome of Acrossocheilus yunnanensis (Cypriniformes: Cyprinidae: Acrossocheilus) and its phylogenetic analysis. Mitochondrial DNA B Resour. 7(10):1764–1765. doi: 10.1080/23802359.2020.1780973.
- Cheng SH, Yan JJ, Liu YL, Lu YM, Zhang Y, Xia MN, Yan YZ. 2015. The complete mitochondrial genome of the Acrossocheilus fasciatus (Cyprinidae, Barbinae). Mitochondrial DNA. 26(6):941–942. doi: 10.3109/19401736.2013.863300.
- Duncker G. 1904. Die Fische der malayischen Halbinsel. Mitteilungen Aus Dem Naturhistorischen Museum in Hamburg. 21:133–207.
- Gong Z, Li X, Chang D, Shen H. 2022. Characterization of the complete mitochondrial genome of garra motuoensis (Cypriniformes: Cyprinidae) and its phylogenetic position within genus Garra. Mitochondrial DNA B Resour. 7(4):663–665. doi: 10.1080/23802359.2022.2064247.
- Gu W, Xu G, Huang T, Wang B. 2020. The complete mitochondrial genome of Neolissochilus benasi (Cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 5(1):463–464. doi: 10.1080/23802359.2019.1703566.
- Guo Y, Wang Y, Yin C, Wang J, Chen Y. 2023. Characterization of the complete mitochondrial genome of Neolissochilus hendersoni (Herre, 1940) (Cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 8(1):133–135. doi: 10.1080/23802359.2022.2163598.
- He J, Zhao C, Guo Y, Zhang H, Zhao B, Chu Z. 2021. Completely mitochondrial genome of Neolissochilus heterostomus. Mitochondrial DNA B Resour. 6(9):2708–2709. doi: 10.1080/23802359.2021.1945966.
- Huang F, Liu M, Ye C, Liu S. 2015. The complete mitochondrial genome sequence of Tor sinensis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 26(5):712–713. doi: 10.3109/19401736.2013.843087.
- Jang-Liaw NH, Chang CH, Tsai CL. 2013. Complete mitogenomes of two Puntius in Taiwan: p. semifasciolatus and P. snyderi (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24(3):228–230. doi: 10.3109/19401736.2012.752472.
- Ju Y-M, Hsu K-C, Yang J-Q, Wu J-H, Li S, Wang W-K, Ding F, Li J, Lin H-D. 2018. Mitochondrial diversity and phylogeography of Acrossocheilus paradoxus (Teleostei: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 29(8):1194–1202. doi: 10.1080/24701394.2018.1431227.
- Khaironizam M, Akaria-Ismail M, Armbruster JW. 2015. Cyprinid fishes of the genus neolissochilus in peninsular malaysia. Zootaxa. 3962(1):139–157. doi: 10.11646/zootaxa.3962.1.7.
- Khare P, Mohindra V, Barman AS, Singh RK, Lal KK. 2014. Molecular evidence to reconcile taxonomic instability in mahseer species (Pisces: Cyprinidae) of India. Org Divers Evol. 14(3):307–326. doi: 10.1007/s13127-014-0172-8.
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Liu XX, Song XL, Li HL, Yuan LY. 2018. The complete mitogenome and phylogenetic analysis of Acrossocheilus beijiangensis (Osteichthyes: Cyprinidae). Mitochondrial DNA B Resour. 3(1):24–25. doi: 10.1080/23802359.2017.1413314.
- Pan T, Jiang H, Hu Y, Ling J, Duan G. 2019. The complete mitochondrial genome of Acrossocheilus wenchowensis (Cyprinidae, Barbinae) from Xinanjiang River. Mitochondrial DNA B Resour. 4(2):3159–3160. doi: 10.1080/23802359.2019.1667916.
- Rainboth WJ. 1996. Fishes of the cambodian mekong. Fao Species Identification Field Guide for Fishery Purposes. 265.
- Ren L, Zhang X, Li Y, Shang Y, Chen S, Wang S, Qu Y, Cai J, Guo Y. 2020. Comparative analysis of mitochondrial genomes among the subfamily sarcophaginae (Diptera: Sarcophagidae) and phylogenetic implications. Int J Biol Macromol. 161:214–222. doi: 10.1016/j.ijbiomac.2020.06.043.
- Sarma, D., Mohan, D., Posti, R., Arya, M., & Ganie, P. A. (2022). The mighty mahseers of the genera Tor, Neolissochilus and Naziritor: a review on resource distribution, biology, ecotourism and conservation. Indian J. Fish., 69(4), 146–169. doi: 10.21077/ijf.2022.69.4.125074-20.
- Sati J, Goel C, Kumar R, Ali S, Patiyal RS, Singh VK, Sahoo PK, Barat A. 2014. Complete mitochondrial genome organization of Tor Putitora. Mitochondrial DNA. 25(4):278-279. doi: 10.3109/19401736.2013.800505.
- Scharpf C. 2015. The authorship of neolissochilus soro (Cypriniformes: Cyprinidae): a correction to khaironizam et al.(2015). Zootaxa. 3986(4):499–500. doi: 10.11646/zootaxa.3986.4.10.
- Singh S, Mishra A, Pavan-Kumar A, Bidyasagar Singh S, Radhakrishnan KV, Nagpure NS, Chaudhari A. 2021. Characterization of the complete mitochondrial genome of Neolissochilus hexastichus (McClelland, 1839). Mitochondrial DNA B Resour. 6(9):2461–2463. doi: 10.1080/23802359.2021.1920505.
- Sun CH, Sun PY, Lao YL, Wu T, Zhang YN, Huang Q, Zhang Q. 2023. Mitogenome of a monotypic genus, Oliotius Kottelat, 2013 (Cypriniformes: Cyprinidae): Genomic characterization and phylogenetic position. Gene. 851:147035. doi: 10.1016/j.gene.2022.147035.
- Tan HH, Lim KK. 2004. Inland fishes from the anambas and natuna islands, south china sea, with description of a new species of betta (Teleostei: Osphronemidae). Raffles Bull Zool. 11:107–115.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. doi: 10.1371/journal.pone.0112963.
- Wei Z, Zhao B, Lin X, Yin C, Xu S, Dai X. 2022. The complete mitochondrial genome of Neolissochilus stracheyi (Osteichthyes: Cyprinidae). Mitochondrial DNA B Resour. 7(8):1492–1493. doi: 10.1080/23802359.2022.2107460.
- Xie XY, Huang GF, Li YT, Zhang YT, Chen SX. 2016. Complete mitochondrial genome of Acrossocheilus parallens (Cypriniformes, Barbinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3339–3340. doi: 10.3109/19401736.2015.1018212.
- Yang L, Sado T, Vincent Hirt M, Pasco-Viel E, Arunachalam M, Li J, Wang X, Freyhof J, Saitoh K, Simons AM, et al. 2015. Phylogeny and polyploidy: resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol Phylogenet Evol. 85:97–116. doi: 10.1016/j.ympev.2015.01.014.
- Yuan L, Song X, Ji B, Zhao C, Liu X. 2017. The complete mitogenome and phylogenetic analysis of Acrossocheilus wuyiensis (Osteichthyes Cyprinidae). J Phylogenetics Evol Biol. 05(02):185. doi: 10.4172/2329-9002.1000185.
- Zhao D, Guo Y, Gao Y. 2022. Natural selection drives the evolution of mitogenomes in Acrossocheilus. PLoS One. 17(10):e0276056. doi: 10.1371/journal.pone.0276056.
- Zhou C, Yuan D, Zhu C, Lei L, Zhang C, Zhou J, Gong J, Zhu L, Li B, Wu Q. 2016. The complete mitochondrion genome of the Barbodes hexagonolepis (Cypriniformes, Cyprinidae). Mitochondrial DNA B Resour. 1(1):158–159. doi: 10.1080/23802359.2016.1144110.
- Zhou M-Y, Wang J-J, Ren J-F, Li F, Wu J-X, Zhou J-J, Li J-L, Yang J-Q, Lin H-D. 2023. Historical landscape evolution shaped the phylogeography and population history of the cyprinid fishes of acrossocheilus (Cypriniformes: Cyprinidae) according to mitochondrial DNA in Zhejiang Province, China. Diversity. 15(3):425. doi: 10.3390/d15030425.

